New RNA CRISPR Tool Normalizes Tau Splicing
Quick Links
A new CRISPR tool proves that good things do sometimes come in small packages. Patrick Hsu and colleagues at the Salk Institute in La Jolla, California, identified a family of bacterial RNA-guided, RNA-selective nucleases. Considerably smaller than known Cas nucleases, they efficiently and specifically knocked down mRNAs in human cells. In a first, Hsu and colleagues used the ribonucleases to manipulate mRNA splicing, too. The scientists reversed aberrant tau mRNA splicing in neurons derived from a patient with frontotemporal dementia (FTD), foreshadowing future therapeutic possibilities.
- Researchers discovered a new family of small RNA-targeting CRISPR nucleases.
- These ribonucleases knock down target mRNA in human cells.
- In neurons, an engineered nuclease corrects mis-splicing of tau.
As an extra bonus, the petite ribonucleases are easier to package into viral vectors and introduce into mammalian cells. Hsu’s work appeared in Cell on March 15. A second group, led by David Cheng and David Scott from Arbor Biotechnologies in Cambridge, Massachusetts, independently discovered the same enzyme family, and published their work the same day in Molecular Cell.
Gene Yeo, University of California San Diego, told Alzforum he’s excited to see more RNA-specific CRISPR effectors being discovered. Yeo previously enlisted CRISPR-Cas to destroy repetitive RNAs linked to ALS and Huntington’s disease, but used Cas9, which normally targets DNA (Batra et al., 2017). Yeo said he’s collaborating with Hsu to test the new effectors, and believes their small size may help not only with expression but with reducing immunogenicity, a problem for bacterial proteins in mammalian hosts.
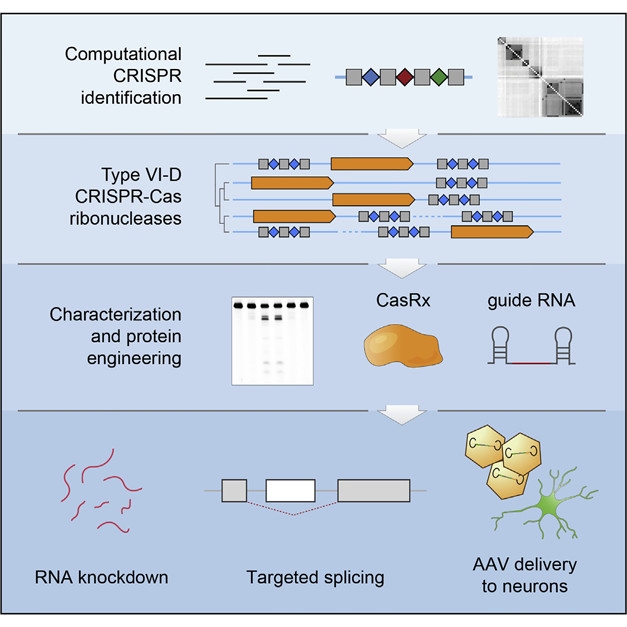
Nice Catch. A computational approach uncovered the type VI-D family of bacterial CRISPR-Cas NA nucleases, which were then engineered for expression in mammalian cells. CasRx and its guide RNA targeted specific transcripts for knockdown and tweaked splicing in human neurons. [Courtesy of Cell, Konermann et al., 2018.]
While much attention has focused on using CRISPR-Cas editing of DNA to treat disease, Hsu sees advantages to targeting RNA. DNA editing permanently alters the genome, and introduces the risk of DNA damage. Changes to DNA tend to be all or nothing; that is good for ablating or replacing a gene wholesale, but not for fine-tuning protein or isoform expression, he told Alzforum. In contrast, targeting dynamic changes in RNA allows for reversible tweaking of both. In addition, an estimated 15 percent of genetic diseases stem from RNA mis-splicing, and would be more amenable to RNA- than DNA-targeted editing approaches.
DNA editing technology has rapidly advanced (Aug 2017 news), but the development of tools to precisely edit RNA has lagged. Only in 2016 did researchers discover bacterial CRISPR-Cas13 nucleases that digest RNA. Last year they reported engineering them to knock down, or make single base changes in, mRNAs in mammalian cells (Abudayyeh et al., 2016; Abudayyeh et al., 2017; Oct 2017 news).
To expand the toolbox, both groups of investigators took the same approach. They scanned prokaryotic sequence databases to uncover novel nucleases related to the previously identified Cas13a and Cas13b enzymes. The new family, which they call Cas13d, comprises the smallest CRISPR effectors described to date. They weigh in at 930 amino acids, compared with 1,120–1,250 for other Cas13s. Both groups extensively characterized the nuclease activity and guide requirements in bacteria, and the Arbor group also identified an accessory protein that boosted Cas13d activity.
Despite their small size, Cas13ds more than matched their cousins in terms of targeted RNA cleavage in human cells. First author Silvana Konermann from Hsu’s group tested seven Cas13d orthologs. They found the best of the lot, from Ruminococcus flavefaciens (CasRx), consistently knocked down levels of 11 different targeted human RNAs by at least 80 percent, with a median of 96 percent reduction. In a head-to-head comparison, shRNA (RNA interference) only reduced expression by 65 percent on average. In Hsu’s hands, CasRx compared favorably with CRISPR-Cas13a and 13b, which decreased target mRNA levels by 80 and 66 percent, respectively. Analysis of whole-cell transcriptomes revealed little off-target mRNA suppression by CasRx, in contrast to shRNA.

Tweaking Tau. Guide RNAs (g1, g2, g3) and a nuclease-dead CasRx (dCasRx) block splicing of exon 10 in the MAPT transcript (left), favoring production of three-repeat tau. In human neurons derived from an FTD patient, guide RNA/dCasRx normalized four-repeat tau levels (right). [Courtesy of Cell, Konermann et al., 2018.]
To modulate splicing, Hsu’s group generated a nuclease-dead mutant (dCasRx) that still bound to RNA, but didn’t cleave it. They suspected that the physical presence of dCasRx on the mRNA would block splicing factors from accessing and processing the message, and that seemed to be the case. When they targeted dCasRx to a splice acceptor site, donor site, or exon in a reporter construct, they did not change overall translation, but did increase exon skipping, with acceptor site targeting being most efficient. “The results suggest that dCasRx may allow investigators to tune isoform expression through use of varying guides,” said Hsu.
Aberrant splicing is implicated in some genetic forms of FTD, where mutations in the MAPT gene for tau elevate levels of a four-repeat (4R) splice isoform relative to the smaller, three-repeat (3R) form. Hsu wondered if dCasRx might normalize tau splicing. He tested this in neurons derived from a patient with a MAPT mutation commonly found in FTD—a C to T change in intron 10 that alters splicing in favor of the 4R form. Before treatment, the neurons had a 4R/3R ratio about three times that of normal control neurons. After adeno-associated virus expression of dCasRx and splice-site targeting guide RNAs, the ratio dropped by about half, bringing it closer to that in control cells.
This is just the beginning for RNA editing, said Hsu. “Splicing and knockdowns are just two of the many things we should be able to do with this platform. When we started with Cas9 DNA editing, the first thing we did was gene knockdown and knock-in. Now we have an entire toolbox for transcriptional control and genome editing in cells. We hope this will be the same for transcriptomics, where you can target any post-transcriptional process, like RNA methylation or editing or translational control, and use these tools to delve deeper in to RNA biology.”
Hsu said he is excited to get CasRx into the hands of more researchers, telling Alzforum the plasmids will be distributed on Addgene. The Arbor Biotechnologies group also said they plan to make their reagents available this way.—Pat McCaffrey
References
News Citations
Mutations Citations
Paper Citations
- Batra R, Nelles DA, Pirie E, Blue SM, Marina RJ, Wang H, Chaim IA, Thomas JD, Zhang N, Nguyen V, Aigner S, Markmiller S, Xia G, Corbett KD, Swanson MS, Yeo GW. Elimination of Toxic Microsatellite Repeat Expansion RNA by RNA-Targeting Cas9. Cell. 2017 Aug 24;170(5):899-912.e10. Epub 2017 Aug 10 PubMed.
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016 Aug 5;353(6299):aaf5573. Epub 2016 Jun 2 PubMed.
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DB, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F. RNA targeting with CRISPR-Cas13. Nature. 2017 Oct 12;550(7675):280-284. Epub 2017 Oct 4 PubMed.
External Citations
Further Reading
No Available Further Reading
Primary Papers
- Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell. 2018 Apr 19;173(3):665-676.e14. Epub 2018 Mar 15 PubMed.
- Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, Scott DA. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol Cell. 2018 Mar 9; PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.