Will The Toxic Aβ Structure Please Stand Up?
Quick Links
A close-up view of short segments of the amyloid-β (Aβ) peptide reveals that they self-associate to form a variety of structures, according to a new study from the lab of David Eisenberg at the University of California, Los Angeles. If these structures accurately model those created by the full-length peptides, then their description may help tease out the details of how amyloid fibrils take on different shapes and clarify whether some configurations are more toxic to neurons than others (see Live Discussion). Such questions have stymied the AD field, as the current understanding of different prefibrillar species, both in terms of their structures and terminology, is confusing at best.
“This study provides insight into the range of structural diversity of the amyloid fibrils formed by a single peptide. The next step will be to determine which structures are important to disease,” said Charles Glabe of the University of California, Irvine, who has collaborated with Eisenberg on a number of studies, but not this latest one published in Proceedings of the National Academy of Science Early Edition. “Anytime we get structural details that can help us understand the mechanism of amyloid formation, I consider it valuable,” said Hilal Lashuel at the Swiss Federal Institute of Technology in Lausanne, who also played no role in the study. “Because we can then leverage this information to achieve the ultimate goals of understanding the structural basis of toxicity and to develop novel approaches to help us diagnose and treat amyloid diseases.”
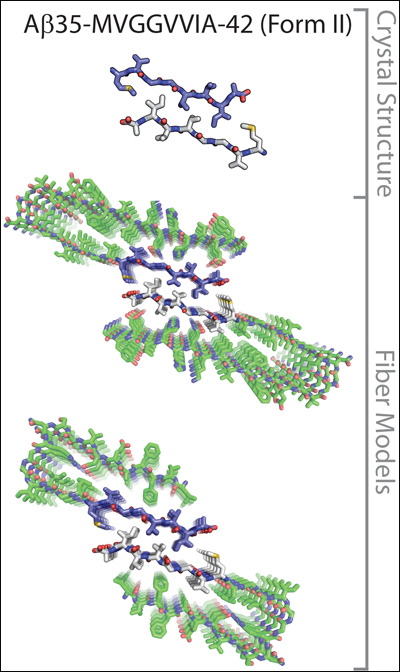
The crystal structures of a seven-amino-acid fragment of the Aβ35-42 peptide model the molecular interactions between two protofilaments of Aβ1-40 (upper) or Aβ1-42 (lower). Image credit: David Eisenberg at UCLA. Reprinted from PNAS Early Edition
It is known that Aβ and other proteins can self-assemble into fibrillary shapes, but piecing together what amyloid fibrils look like at the molecular level has not been an easy task. Researchers have used cryoelectron microscopy and solid-state nuclear magnetic resonance (NMR) imaging to visualize different Aβ aggregates. This has revealed many details of their three-dimensional structures, including the fact that they contain parallel β-sheets (see, e.g., Paravastu et al., 2008). Another method for zooming in on these structures, which can yield even greater detail, is x-ray crystallography, but so far amyloid fibrils and other β-sheet-containing aggregates have proven too large to crystallize.
Eisenberg’s group got around the problem by coaxing small segments, six to seven amino-acids long, of different amyloid-forming peptides into creating three-dimensional crystals less than 1 micrometer in size, which are amenable to x-ray diffraction studies. They first applied the technique to the yeast prion protein Sup35, showing that tiny segments from this protein bound to copies of themselves to create stacked β-sheets (see ARF related news story on Nelson et al., 2005). The amino acid side chains sticking out from each β-strand form interdigitated connections, which resemble the teeth of a zipper and hold the β-sheets in a tight grip. Presumably, in cells, these structures, which Eisenberg called “steric zippers,” allow the amyloid fibers to grow in size as additional β-sheets get stacked on top of each other.
In a follow-up study (Sawaya et al., 2007), Eisenberg described several different steric zipper shapes formed by various amyloid-forming peptide segments, including ones from Aβ and the tau protein. The latest study in this series extends these observations, focusing exclusively on protein segments derived from Aβ. In the current paper, Eisenberg’s group reported eight new crystal structures of Aβ segments. When added to the three determined previously, these 11 structures reveal that identical Aβ segments can interact in different ways to give rise to 13 different steric zipper arrangements. The authors then overlaid their crystal structures on lower-resolution structures of amyloid fibrils obtained by NMR to generate several models of protofilament, the precursors to fibrils, which reveal the molecular interactions between peptide strands. The models suggest that Aβ can form a wide range of possible fibril structures, or polymorphs. “It is quite surprising that a single peptide can take on all these different forms,” said Glabe.
The notion that Aβ molecules can form different types of fibril structures is not new. Solid-state NMR and electron microscopy measurements on full-length Aβ fibrils revealed several such forms (Petkova et al., 2005; Williams et al., 2005; Kodali et al., 2007; Paravastu et al., 2009). “We have published specific structural models for two polymorphs that give distinct NMR spectra and have distinct appearances in electron microscope images,” wrote Robert Tycko of the National Institutes of Health in Bethesda, Maryland in an e-mail to ARF (see full comment below). “Some of the models in the Eisenberg paper resemble our solid-state NMR-based models, while others are quite different.” In addition, conformation-specific antibodies have identified different types of prefibrillar amyloid oligomers (see ARF related news story).
This latest study adds to the array of polymorphs that have been detected. By using x-ray crystallography, Eisenberg and colleagues were able to describe the interactions between β-sheets in greater detail than before. On the other hand, the drawback of Eisenberg’s technique is that it is based on short peptide fragments. “These models indicate the potential diversity in fibril structures, but we don’t know if it is fully utilized in the full-length peptide,” said Glabe.
One application of this knowledge is the development of drugs that prevent fibrils and other β-sheet aggregates from forming. “Until now, we have not had atomic-level information to design such inhibitors,” Eisenberg said. His group has already started using a structure-based approach to find a tau inhibitor (see ARF related news story on Sievers et al., 2011).
The million-dollar question is whether there is a particular molecular structure of Aβ fibrils that characterizes AD in the brain. “That is what everyone would like to know,” said Eisenberg. Researchers have long speculated that if Aβ can form different structures, then they would have different biological effects, said Lashuel. Several groups are chasing this idea. Other scientists, however, suspect the process of aggregation itself, more than any one particular species, is at fault in AD. Future experiments will provide more definitive information on both fronts.—Laura Bonetta
References
News Citations
- Making Heads and Tails of Prion Amyloid
- Oligomers Abound, But Where’s the Seed?
- Structure-Based Approach Yields Tau Inhibitors
Paper Citations
- Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2008 Nov 25;105(47):18349-54. PubMed.
- Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005 Jun 9;435(7043):773-8. PubMed.
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen AØ, Riekel C, Eisenberg D. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007 May 24;447(7143):453-7. PubMed.
- Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005 Jan 14;307(5707):262-5. PubMed.
- Williams AD, Sega M, Chen M, Kheterpal I, Geva M, Berthelier V, Kaleta DT, Cook KD, Wetzel R. Structural properties of Abeta protofibrils stabilized by a small molecule. Proc Natl Acad Sci U S A. 2005 May 17;102(20):7115-20. PubMed.
- Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr Opin Struct Biol. 2007 Feb;17(1):48-57. PubMed.
- Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. Seeded growth of beta-amyloid fibrils from Alzheimer's brain-derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci U S A. 2009 May 5;106(18):7443-8. PubMed.
- Sievers SA, Karanicolas J, Chang HW, Zhao A, Jiang L, Zirafi O, Stevens JT, Münch J, Baker D, Eisenberg D. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature. 2011 Jul 7;475(7354):96-100. PubMed.
Other Citations
Further Reading
Papers
- Basi GS, Feinberg H, Oshidari F, Anderson J, Barbour R, Baker J, Comery TA, Diep L, Gill D, Johnson-Wood K, Goel A, Grantcharova K, Lee M, Li J, Partridge A, Griswold-Prenner I, Piot N, Walker D, Widom A, Pangalos MN, Seubert P, Jacobsen JS, Schenk D, Weis WI. Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease. J Biol Chem. 2010 Jan 29;285(5):3417-27. PubMed.
Primary Papers
- Colletier JP, Laganowsky A, Landau M, Zhao M, Soriaga AB, Goldschmidt L, Flot D, Cascio D, Sawaya MR, Eisenberg D. Molecular basis for amyloid-beta polymorphism. Proc Natl Acad Sci U S A. 2011 Oct 11;108(41):16938-43. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
NIDDK, NIH
This paper from the Eisenberg group suggests several molecular structural models for fibrils formed by full-length β amyloid peptides, based on their crystal structures of segments of the full-length peptide that reveal the details of intermolecular interactions. Using solid-state NMR and electron microscopy measurements on full-length β amyloid fibrils, my group showed earlier that β amyloid can form multiple distinct fibril structures (i.e., multiple polymorphs), and we have published specific structural models for two polymorphs that give distinct NMR spectra and have distinct appearances in electron microscope images. These experiments were done on fibrils that were prepared in vitro from synthetic β amyloid peptide. Some of the models in the Eisenberg paper resemble our solid-state NMR-based models, while others are quite different. The total number of fibril structures that β amyloid can form has not been determined, but is at least five, based on results from Ron Wetzel's lab and my lab. The new fibril structures suggested in the Eisenberg paper are plausible, and may be identified in future studies of other full-length β amyloid fibril polymorphs.
We suggested in 2005 (Petkova et al., 2005) that β amyloid fibril polymorphism may have biomedical significance, with certain polymorphs effectively being more neurotoxic than others. We subsequently obtained evidence that fibrils in AD brain tissue have molecular structures that are different from our published structural models (Paravastu et al., 2009). We do not yet know whether there is a connection between variations in β amyloid fibril structure and the development of AD, but this remains a viable and interesting hypothesis that we are pursuing. The possibility exists that the structural models suggested in the Eisenberg paper do correspond to fibrils that occur in AD. Future experiments will provide more definitive information.
References:
Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005 Jan 14;307(5707):262-5. PubMed.
Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. Seeded growth of beta-amyloid fibrils from Alzheimer's brain-derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci U S A. 2009 May 5;106(18):7443-8. PubMed.
Make a Comment
To make a comment you must login or register.