Aggregation-Prone Gene Expression Signature Mapped in Brain
Quick Links
Some areas of the brain seem to lay out a welcome mat for Alzheimer's, while others remain impervious to the disease throughout its course. Now, researchers report that this regional vulnerability has a signature. According to a brain-wide transcriptional analysis published in the August 10 Science Advances, areas of the brain hit earliest and hardest by AD highly express genes associated with protein aggregation. Those same areas turn down genes known to break up protein clogs. Although the researchers, led by Michele Vendruscolo at the University of Cambridge, U.K., await proteomic data to clinch the connection between gene expression and protein levels, the study provides strong support for the hypothesis that tissue vulnerability dictates when and where neurodegenerative disease strikes in the brain.
“The biology underlying the specific hierarchical vulnerability patterns has been a mystery for years, and this work has the promise of providing a framework to understand it better,” commented Bradley Hyman of Massachusetts General Hospital in Charlestown, who was not involved in the work. “This is a ‘must read’ for everyone interested in the balance between protein homeostasis and protein aggregation biology in neurodegenerative syndromes.”
Why different neurodegenerative diseases attack specific parts of the brain remains unclear. In AD, tangles first appear in the entorhinal cortex (EC), then the hippocampus, and eventually other regions of the cortex succumb (see Braak and Braak, 1991). The reasons for the EC’s vulnerability and exactly how pathology spreads from there to other regions are heavily investigated questions in the field. Some propose that misfolded proteins such as Aβ and tau spread along connected neural networks and, like prions, corrupt normal versions of the proteins along the way (see Walker and Jucker, 2013). Instead of (or in addition to) such proteopathic spread, differential vulnerability between brain regions could explain the spatiotemporal progression of pathology (see Miller et al., 2013). Vendruscolo and colleagues proposed that both could be true—perhaps traveling misfolded proteins only settle down in the most vulnerable neighboring regions. They hypothesized that each region’s vulnerability to AD pathology could be explained by how well it prevented the accumulation of aggregated proteins, which would be reflected in the relative expression of certain genes.
First author Rosie Freer and colleagues set out to test this hypothesis by tapping into gene-expression data from the Allen Brain Atlas. The researchers accessed genome-wide transcriptome data from 500 regions in six brains from healthy people, aged 24 to 57. Superimposed on these 500 regions, the researchers cast Braak staging criteria, which define each area’s vulnerability to pathology based on when (and if) it develops neurofibrillary tau tangles as AD progresses.
They first looked at expression levels of genes encoding proteins known to co-aggregate with Aβ or tau in plaques and tangles, respectively, and found that compared to so-called “non-Braak” regions (which never develop AD pathology), Braak regions I to III (where tau pathology first pops up) expressed higher levels of these genes. To quantify these differences, the researchers gave each gene “Δ scores,” which reflect the difference in that expression between a given Braak region and all non-Braak regions. While not every gene encoding a known co-aggregating protein had a positive Δ score for Braak regions I to III, a majority of them, including APP, tau, tubulin, dynamin, and calcium calmodulin, were highly expressed in these regions. Interestingly, they also found that proteins co-mingling in Lewy bodies that characterize Parkinson’s disease were more highly expressed in the substantia nigra pars compacta, which is the primary site of vulnerability for that disease.
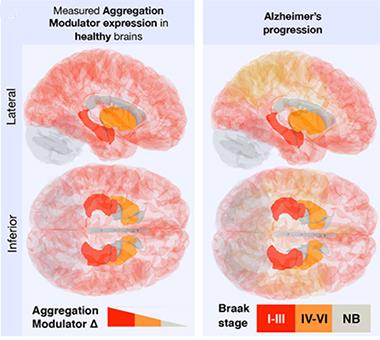
Opening the Door to AD?
Vulnerability to AD as measured by Braak staging (right) correlates with the expression of aggregation-promoting genes (left). [Image courtesy of Freer et al., Science Advances, 2016.]
The researchers next asked whether the expression of factors known to influence the aggregation of Aβ and tau—including chaperones, proteases, and other post-translational modifiers—would be turned up or down in regions affected by AD. They compiled a list of such proteins, checked their expression levels, and assigned Δ scores. They found that genes encoding proteins known to promote aggregation were also more active in early Braak regions, while those known to prevent aggregation were less active.
Some of the regions sectioned off in the Allen Brain Atlas did not align perfectly with regions cordoned off in the Braak staging map. To get a tighter association between gene expression and staging, the researchers next limited their comparisons to regions that aligned perfectly between the two maps. They found that the expression patterns of the aggregation modulators recapitulated the staging of the disease.

AD Waiting to Happen? In areas of the brain where AD strikes earliest, genes associated with aggregation promotion (ellipses) tended to be overly active, while those that control aggregation (rectangles) are less active. [Image courtesy of Freer et al., Science Advances, 2016.]
The researchers found evidence for the importance of the immune system in AD vulnerability as well. Expression of genes associated with inflammation were elevated in AD-vulnerable tissues, while those involved in autoimmune responses were lower. While Vendruscolo would not speculate on the meaning of these results, he said they are consistent with the idea that inflammation mediates the disease process.
Drilling down from regional vulnerability, could different cell types also harbor gene expression signatures that predispose them to AD pathology? To find out, the researchers compared single-cell mRNA sequencing data from neurons, astrocytes, microglia, and endothelial cells. They found that neurons expressed the highest levels of Aβ, tau, and aggregation promoters, but the lowest levels of aggregation protectors. This indicated that compared with other cell types, neurons may provide a welcoming environment for AD pathology.
“The notion that there are differences between brain regions regarding their ‘permissiveness’ to protein-misfolding disease has been discussed for decades, but strong supporting data have been lacking,” commented Patrik Brundin of the Van Andel Research Institute in Grand Rapids, Michigan. “Notably, this study highlights the possibility that there exist region-specific differences both in the levels of proteins that promote or protect against protein aggregation, as well as in the levels of proteins engaged in immune system signaling.” However, Brundin noted that the most burning question—what actually triggers the disease in vulnerable regions—is still unanswered (see full comment below).
Vendruscolo acknowledged that one major caveat with the findings is that they measure expression of the RNA rather than protein, and sometimes the two are not strongly correlated. However, he said the researchers partially addressed this issue by averaging Δ scores over multiple genes, and also corroborated their findings with protein-level data available for mouse brains. Vendruscolo plans to repeat their analysis when regional and cell-specific proteomic data becomes available for the human brain.
Vendruscolo emphasized that while the findings are only a first step, they support the idea that protein aggregation is at the heart of tissue vulnerability to AD pathology in the brain. He added that the findings are consistent with the concept of proteopathic protein spread as well. “Aggregated proteins may spread to many neighboring regions, but only the vulnerable regions will get disease,” he said.
Brundin agreed. “Essentially, the findings imply that some regions are more susceptible to develop neuropathology once proteopathic seeds are delivered from other brain areas, and that the same regions might be less effective at clearing aggregates via the immune system.”—Jessica Shugart
References
Paper Citations
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239-59. PubMed.
- Jucker M, Walker LC. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. 2013 Sep 5;501(7465):45-51. PubMed.
- Miller JA, Woltjer RL, Goodenbour JM, Horvath S, Geschwind DH. Genes and pathways underlying regional and cell type changes in Alzheimer's disease. Genome Med. 2013 May 25;5(5):48. PubMed.
Further Reading
Primary Papers
- Freer R, Sormanni P, Vecchi G, Ciryam P, Dobson CM, Vendruscolo M. A protein homeostasis signature in healthy brains recapitulates tissue vulnerability to Alzheimer's disease. Sci Adv. 2016 Aug;2(8):e1600947. Epub 2016 Aug 10 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
I think this is a really interesting advance. The biology underlying the specific hierarchical vulnerability patterns has been a mystery for years, and this work has the promise of providing a framework to understand it better. While clearly the patterns of connections play some role, it has always been obvious that specific populations of neurons have a certain predilection toward neurodegeneration in different neurological diseases, likely either superimposed on constraints placed by connectivity, or perhaps even in some ways coordinated with it. This is a “must read” for everyone interested in the balance between protein homeostasis and protein aggregation biology in neurodegenerative syndromes.
University of Melbourne
This seems like a good systems biology approach to understanding regional vulnerability in protein-aggregating neurodegenerative diseases. The “promoters” and “protectors” selected for this initial analysis are a relatively small series, and no doubt will be supplemented by others as we learn more about the underlying mechanisms that drive aggregation.
The current reliance only on Braak staging of regional areas might also be enhanced in future work by consideration of in-vivo molecular imaging with Aβ and tau PET studies, which are probably more informative than the classic histological approaches.
Nevertheless, the identification of a neuronal protein homeostasis signature points strongly to an innate selective vulnerability of certain brain areas, which takes us back to one of the very first neuropathological concepts of “pathoclisis.”
Associate Director of the Van Andel Research Institute, and Director of the Center for Neurodegenerative Science
Freer and collaborators’ findings are very interesting indeed. The notion that there are differences between brain regions regarding their “permissiveness” to protein misfolding disease has been discussed for decades, but strong supporting data have been lacking. Notably, this study highlights the possibility that there exist region-specific differences both in the levels of proteins that promote or protect against protein aggregation, as well as in the levels of proteins engaged in immune system signaling.
Taken together, the authors suggest that this makes certain brain regions more or less vulnerable to protein misfolding diseases. It is important to stress that the region-specific differences were observed in postmortem brains from normal subjects, and that there is no evidence that levels of “promotors” or “protectors” in a given individual reveal anything about whether he/she is more or less likely to develop disease.
Further, the authors agree that the findings tell us little about perhaps the most burning issue, namely what the triggers of the diseases are. Importantly, the novel findings remain consistent with the notion that protein aggregates can spread along neural pathways in a prion-like fashion.
Essentially, the findings imply that some regions are more susceptible to developing neuropathology once proteopathic seeds are delivered from other brain areas, and that the same regions might be less effective at clearing aggregates via the immune system. Future studies might help clarify why these region-specific differences have evolved, and how they are coupled to differences in the normal functions of the different brain regions.
RIKEN Center for Brain Science
This is an innovative work, which, at least in part, explains the region-specific vulnerability of brain to neurodegenerative processes. The presented scheme is beautiful. I do however have a concern. Systems biology relies on the presumption that all the molecular cause-and-effect relationships described thus far are essentially correct, but this may not always be true. For instance, we still do not know whether phosphorylation is a cause or consequence of tauopathy, questioning the role of GSK3 in AD pathogenesis.
Make a Comment
To make a comment you must login or register.