Widely Used Tau Seeding Assay Challenged
Quick Links
Many research groups study tau misfolding and propagation using in vitro models, but interpreting findings from artificial systems can be dicey. In a preprint on bioRxiv, researchers led by Eckhard Mandelkow at the German Center for Neurodegenerative Diseases, Bonn, question whether a widely used assay of tau propagation truly detects transmission of a toxic conformation.
- A fluorescent tag prevents tau fragments from forming paired helical filaments.
- This implies tau inclusions in a common propagation assay may not be PHFs.
- The finding casts doubt on whether the assay detects prion-like propagation.
In this assay, researchers add extracts from Alzheimer’s brain to cultured cells that contain fluorescently labeled tau fragments. When seeded, these fragments aggregate and light up via fluorescence resonance energy transfer (FRET), suggesting a prion-like spread of misfolded tau from the outside of the cell to its inside. However, Mandelkow and colleagues found that the bulky fluorescent label on these tau fragments creates a steric hindrance that prevents them from forming paired helical filaments (PHFs), the building blocks of tangles. Instead, the labeled fragments coalesce into amorphous clumps.
The findings suggest that this assay cannot detect prion-like propagation. This would throw open the possibility that something else in the brain extract might be responsible for inducing tau aggregation, the authors argue. “Factors other than tau should be considered as the [extracellular] agents propagating tau pathology,” Mandelkow wrote to Alzforum.
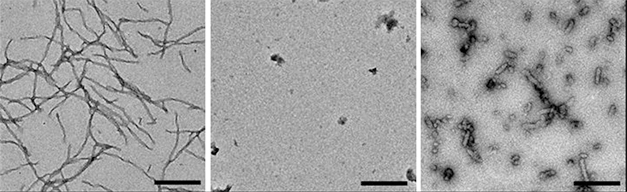
Packing Interference. Unlabeled aggregation-prone tau fragments form long fibrils (left). Those with an N-terminal fluorescent tag do not (middle), and those with a C-terminal tag form short, aberrant fibrils (right). [Courtesy of Kaniyappan et al., bioRxiv.]
Other scientists disagree with this conclusion. They accept the finding that these fluorescently labeled tau fragments cannot form PHFs, but still believe the cellular assay flags the presence of pathological tau in the brain extract. “I believe these types of cellular aggregation assays are useful and powerful tools to detect seeding-competent assemblies from human or animal brain,” Wouter Peelaerts at the Van Andel Research Institute in Grand Rapids, Michigan, wrote to Alzforum. To Brad Hyman at Massachusetts General Hospital in Charlestown, the data highlight the need to pay attention to the limitations of cellular assays. “There was never any question that the conformational structure of the FRET-based bioreporter would be the same as tau aggregates in the brain … Like most models, its utility depends heavily on understanding its strengths and weaknesses,” he wrote (full comment below). Hyman uses the assay in his own work.
The FRET assay was originally developed by Marc Diamond, now at the University of Texas Southwestern Medical Center in Dallas. He engineered HEK293 cells to express aggregation-prone fragments of tau that are tagged with either a donor (cyan) or acceptor (yellow) fluorescent molecule. When researchers add tau fibrils to the culture medium, these labeled tau pieces come together and fluoresce (Oct 2014 news). The assay has since been adopted by many other labs as a way to measure tau’s pathological activity.
To take a closer look at the structure of these fluorescent tau aggregates, joint first authors Senthilvelrajan Kaniyappan and Katharina Tepper in Mandelkow’s group created tagged tau constructs similar to those used in the FRET assay. They took the short repeat domain of tau containing the pro-aggregant deletion mutation ΔK280, and fused it with a GFP tag at either the N- or C-terminus. Then they incubated these constructs with the nucleating agent heparin in cell-free solution and analyzed the results by UV light scattering. Unlabeled tau fragments aggregated readily, while C-terminally tagged tau formed only a third as many aggregates, and N-terminally tagged tau formed none. Electron microscopy revealed that C-terminally labeled tau aggregated into short fibrils, about twice as thick as those generated by unlabeled tau, with a distinct appearance from PHFs. Meanwhile, N-labeled tau clumped up randomly (see image above).
Additional structural studies added evidence that labeled tau assembles differently than unlabeled. Scanning transmission electron microscopy showed that unlabeled tau fragments formed filaments with a mass-per-length ratio of 4.4 molecules per nanometer, in good agreement with the expected value of 4.3 for PHFs. Labeled tau fragments, on the other hand, formed aggregates with a mass-per-length value around 2. “These values are clearly incompatible with a PHF-like packing of molecules,” the authors wrote. Mandelkow noted that GFP has a diameter of about 3 nm, while the rungs of PHFs are about half a nanometer apart, so there is no room to fit a GFP tag on every rung. The size of the GFP tag prevents close packing of the tau protein, Mandelkow concluded.
In a comment on bioRxiv, Diamond and colleagues question whether the findings from these cell-free studies apply to their cellular assay. Diamond noted several technical differences, chief among them his group’s use of a longer linker sequence, 21 amino acids instead of 13, to attach the fluorescent tag. That would allow GFP to float farther from the fibril core and give it more room to pack. The FRET assay uses a different tau mutation, P301S, and different fluorescent tags, CFP and YFP. Diamond also noted that PHFs are highly polymorphic, with diameters that can vary by more than 2.6-fold, and pointed to a prior study that found evidence for tau aggregates assuming a β-sheet structure in this cellular assay (Barghorn and Mandelkow, 2002; Wegmann et al., 2010; Sanders et al., 2014).
Mandelkow disputes the idea that the GFP linker would make a significant difference, noting that because amino acid strands coil up in solution, the difference in length would be only about one nanometer, not enough to allow the GFP molecules to pack together.
Lary Walker at Emory University, Atlanta, said the Mandelkow group’s conclusions were reasonable within the context of their experimental conditions, and noted that the cellular environment complicates things. “To settle the issue, it would be useful to run controlled comparisons of technical differences such as the linker length in both paradigms,” Walker wrote to Alzforum (full comment below).
Peelaerts cautioned that perhaps none of these in vitro systems reflect the behavior of tau in the brain. “PHFs are just one part of a bigger puzzle. Aggregated tau exists in many conformations, which are dynamic and driven by the equilibrium between the cellular environment and the protein itself. Comparing in vitro assembled seeds with more physiological conditions is therefore always a difficult exercise,” he wrote.
Beyond the structural issue, the scientists also disagreed on the broader interpretation of a positive FRET signal in this assay, and whether that indicates the presence of misfolded tau in the brain extract. Ben Wolozin at Boston University concurred with Hyman and Peelaerts that the assay responds to misfolded tau. “Multiple published studies show that the FRET-sensor lines reliably detect the presence of aggregation-competent tau in brain tissues,” Wolozin wrote. He noted that his company, Aquinnah Pharmaceuticals, has found good concordance between a positive signal in this assay and detection of tau aggregates in the same brain extract using biochemistry or immunohistochemistry. Aquinnah searches for ways to eliminate stress granules, which are associated with Alzheimer’s disease and amyotrophic lateral sclerosis.
For his part, Mandelkow believes the intracellular tau deposits seen in the FRET assay may represent a response to cellular stress or inflammatory stimuli, rather than to aggregated tau in the extract. He noted that tau in primary mouse neurons can be induced to aggregate simply by exposure to activated microglia, or treatment with the proinflammatory cytokine TNFα (Gorlovoy et al., 2009). In addition, a recent study implicated activated microglia in promoting tangles in a tau mouse model (Nov 2019 news). Perhaps inflammation, rather than prion-like propagation, stokes the spread of misfolded tau through brain, Mandelkow suggested.—Madolyn Bowman Rogers
References
News Citations
- Cellular Biosensor Detects Tau Seeds Long Before They Sprout Pathology
- Microglia Inflammasome Stokes Tau Phosphorylation, Tangles
Paper Citations
- Barghorn S, Mandelkow E. Toward a unified scheme for the aggregation of tau into Alzheimer paired helical filaments. Biochemistry. 2002 Dec 17;41(50):14885-96. PubMed.
- Wegmann S, Jung YJ, Chinnathambi S, Mandelkow EM, Mandelkow E, Muller DJ. Human Tau isoforms assemble into ribbon-like fibrils that display polymorphic structure and stability. J Biol Chem. 2010 Aug 27;285(35):27302-13. PubMed.
- Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirbaha H, Li A, Barker SJ, Foley AC, Thorpe JR, Serpell LC, Miller TM, Grinberg LT, Seeley WW, Diamond MI. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron. 2014 Jun 18;82(6):1271-88. Epub 2014 May 22 PubMed.
- Gorlovoy P, Larionov S, Pham TT, Neumann H. Accumulation of tau induced in neurites by microglial proinflammatory mediators. FASEB J. 2009 Aug;23(8):2502-13. PubMed.
External Citations
Further Reading
News
- Antibodies Stop Toxic Tau in Its Extracellular Tracks
- To Block Tau’s Proteopathic Spread, Antibody Must Attack its Mid-Region
- Connectivity, Not Proximity, Predicts Tau Spread
- Do Immune Cells Promote the Spread of α-Synuclein Pathology?
- Human Tau Strains Propagate Faithfully in Wild-Type Mice
- Tau Filaments from the Alzheimer’s Brain Revealed at Atomic Resolution
- Does Tau’s Third Repeat Propagate Misfolding in Vivo?
Primary Papers
- Kaniyappan S, Tepper K, Biernat J, Chandupatla RR, Hübschmann S, Irsen S, Bicher S, Klatt C, Mandelkow EM, Mandelkow E. FRET-based tau seeding assay does not represent prion-like templated assembly of tau fibers. bioRxiv March 25, 2020. BioRxiv.
- Kaniyappan S, Tepper K, Biernat J, Chandupatla RR, Hübschmann S, Irsen S, Bicher S, Klatt C, Mandelkow EM, Mandelkow E. FRET-based Tau seeding assay does not represent prion-like templated assembly of Tau filaments. Mol Neurodegener. 2020 Jul 16;15(1):39. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
Boston University School of Medicine
Understanding mechanisms of tau aggregation continues to be an important avenue of ongoing research. A challenge we all face in modeling tau aggregation as it occurs in AD and ADRD is that the milieu of the neuron is much more complicated than that occurring in vitro or even in cell lines. This differential complexity between neurons and in vitro milieus raises strong challenges for modeling the actual process that gives rise to tau pathology and toxic tau oligomers.
The manuscript by Kaniyappan et al. comes from the Mandelkow laboratory and highlights important differences between tau aggregation occurring in the brain and tau aggregation occurring in FRET-based biosensor lines, such as those developed by the Diamond group. The Mandelkow team shows that the presence of fluorescent proteins in the chimeric recombinant tau constructs used in the FRET-based biosensor lines gives rise to tau fibrils that are structurally very different than native tau aggregates occurring in vitro or in the brain. This is perhaps not surprising, because the fluorescent proteins are larger than the small tau peptides (such as the K18 peptide) that drive tau aggregation in the FRET-based biosensor lines. Thus, the fluorescent proteins take up space and change the structure of the resulting filament.
The differences in filament structure pose important restrictions on how one can interpret results from such biosensor lines. The Mandelkow team correctly points out that one cannot use these biosensor lines as the basis of structural studies of tau filament formation.
However, these biosensor lines have other uses that remain viable, even if the resulting tau filaments do not reflect those occurring in AD and ADRD. One important use is as a sensitive assay to detect the presence of aggregation-competent tau in brain tissues (e.g., from transgenic mice) or in experiments using other cultured cells. The FRET-based biosensors robustly form aggregates when seeded with aggregation-competent tau. In this scenario, the biosensor line is used to detect a particular biochemical species, and the nature of the resulting signal is less important than the sensitivity and specificity of detection. Using an ELISA assay presents a good analogy. The signal from the ELISA assay reflects the presence of tau oligomers or aggregates, but the actual signal itself is structurally very different than the tau oligomer/aggregate.
A second use of the biosensor lines is to detect conformational differences in tau aggregates. In this scenario, the structure of the resulting biosensor aggregate is less important than the pattern of accumulation of tau aggregates, which tends to selectively reflect the initiating tau aggregate conformations.
Emory University
Having speed-read the tau controversy, I would conclude that the findings of the Mandelkow group are reasonable within the context of their experimental conditions. The problem is one of comparing apples and oranges, as pointed out by Diamond et al. The Diamond model of in vivo (cell culture) seeding is well validated in multiple labs.
To settle the issue, it would be useful to run controlled comparisons of technical differences, such as the linker length for the fluorescent molecules, in the Mandelkow paradigm. It would also be useful if the Mandelkow group could perform cell culture experiments using the Diamond model. I really like the degree of control you can get with in vitro paradigms such as the Mandelkows’. These are quite useful in finely dissecting molecular mechanisms, but the cellular environment can complicate things considerably.
There really never was a question that the conformational structure of the FRET-based bioreporter would be the same as tau aggregates in the brain–the former is short and has large fluorescent proteins attached, the latter is full-length and has innumerable post-translational modifications. Indeed, even recombinant tau, if aggregated with heparin, forms a very different structure than PHF.
The key question is whether the FRET reporter assay allows one to examine biological processes that are of import to the disease. The work of Diamond and numerous other labs suggests that the answer is “yes” in many circumstances, although the Mandelkow data elegantly shows that the answer is “no” in terms of detailed conformational studies intended to model the seed that was introduced.
Thus, like most models, its utility depends heavily on understanding its strengths and weaknesses, and seeing how they impact the experimental question at hand.
Sylics
This is an interesting study as it provides valuable information on the nature of the tau aggregates that underlie the fluorescent signal one detects in this seeding assay. In addition, it provides a very useful characterization of how fluorophores linked to different tau constructs influence aggregation. The results of this study suggest that a deeper characterization of the different tau seeding assays might be worthwhile. Perhaps assays that lead to accumulation of tau fibrils are more sensitive biosensors (e.g., generate more signal)? It might also be necessary for certain studies to use seeding-based biosensor assays that generate fibrils that more closely resemble those of tauopathy patients, for example to study how tau seeding impacts the cell or induces tau propagation to other cells.
It should also be noted that the main conclusion of this article might only apply to biosensor assays with the repeat domain of tau linked to the fluorophore. The data in the manuscript show that fibrils do form when the fluorophore is linked to the N-terminal or C-terminal of full-length tau. It remains to be determined if aggregates composed of full-length tau linked to fluorophores—induced by human brain-derived seeds—more closely resemble the fibrils found in the brains of tauopathy patients.
In addition to the previously posted comments to this article, it is worth noting that other versions of this assay use labelled antibodies to stain the tau aggregates after seeding to obtain a FRET signal. The observed characteristics of this assay are similar to the original tau biosensor assay (Courade et al, 2018).
References:
Courade JP, Angers R, Mairet-Coello G, Pacico N, Tyson K, Lightwood D, Munro R, McMillan D, Griffin R, Baker T, Starkie D, Nan R, Westwood M, Mushikiwabo ML, Jung S, Odede G, Sweeney B, Popplewell A, Burgess G, Downey P, Citron M. Epitope determines efficacy of therapeutic anti-Tau antibodies in a functional assay with human Alzheimer Tau. Acta Neuropathol. 2018 Nov;136(5):729-745. Epub 2018 Sep 20 PubMed.
Roche
DZNE German Center of Neurdeg. Diseases
DZNE (German Center for Neurodegenerative Diseases)
In our paper, now in press at Mol. Neurodegeneration, we argue that the repeat domain of Tau, when coupled to GFP, cannot form amyloid-like Tau filaments because of steric hindrance. GFP is too big to fit onto the tight cross-β structure of an amyloid fiber; analogous observations have been made with other amyloid proteins. This implies that the local foci, observed in a Diamond-type sensor cell expressing FRET pairs of TauRD-XFP, are not caused by amyloid Tau fibers. This contradicts the hypothesis that the propagation of Tau pathology is based on a prion-like propagation of Tau assembly. However, it would be compatible with a local accumulation of TauRD-XFP in a non-amyloid state.
Several colleagues have made the counterargument that the sensor cell reaction is a reliable indicator of some pathological property in the Tau preparations used to trigger the FRET reaction (e.g., extracts from AD brains or transgenic mouse models). We agree (and say so in the paper), but this only means that the sensor cell is a useful diagnostic tool.
By analogy, in ALS research arsenite is used to induce local foci of XFP-labeled FUS in the cytoplasm, but this does not mean that arsenite templates the amyloid assembly of FUS; rather, it works indirectly by triggering stress granules containing FUS-XFP.
The implication of our argument is that not Tau itself, but some other Tau-related agent may be the trigger for the FRET reaction. In support of this, the same type of FRET reaction can be triggered by non-Tau agents, e.g. pro-inflammatory cytokines; therefore the trigger is not unique.
In essence, we argue that one should strictly distinguish between the spreading of Tau pathology, which is generally accepted by Braak staging, and the spreading of Tau protein by templated assembly, which is a matter of debate, and generally adopt a less Tau-centric view on the origins of Tau pathology. After all, only 25 percent of AD diversity can be ascribed to Tau, as shown by Dujardin et al., 2020.
References:
Dujardin S, Commins C, Lathuiliere A, Beerepoot P, Fernandes AR, Kamath TV, De Los Santos MB, Klickstein N, Corjuc DL, Corjuc BT, Dooley PM, Viode A, Oakley DH, Moore BD, Mullin K, Jean-Gilles D, Clark R, Atchison K, Moore R, Chibnik LB, Tanzi RE, Frosch MP, Serrano-Pozo A, Elwood F, Steen JA, Kennedy ME, Hyman BT. Tau molecular diversity contributes to clinical heterogeneity in Alzheimer's disease. Nat Med. 2020 Aug;26(8):1256-1263. Epub 2020 Jun 22 PubMed. Correction.
Make a Comment
To make a comment you must login or register.