SLICK Science—Imaging Advances May Aid Brain Research
Quick Links
Peering through the dense thicket of axons and dendrites to figure out what individual neurons are up to in brain tissue is no mean feat. It requires both probing the function of individual neurons and visualizing them at the same time. In the May 4 Nature Neuroscience, researchers led by Guoping Feng at Duke University, Durham, North Carolina, introduce a way to do both. Called SLICK—for single-neuron labeling with inducible Cre-mediated knockout—the method labels individual neurons and knocks out targeted genes at the same time. The researchers used SLICK to study neuromuscular synapses. However, “it could easily be adapted to understand the mechanisms that regulate neuronal stability in the CNS, and this could have significant implications in aging research,” said Feng in an interview with ARF. In other imaging news this week, researchers led by Roger Tsien at the University of San Diego, California, report a new screen for photostable fluorescent probes. With this screen, the researchers developed several new fluorophores that work just as well as green fluorescent protein but have their peak emission at different parts of the visual spectrum. The report appears in the May 4 Nature Methods online.
The SLICK method relies on a genetic construct that drives transcription of both the Cre recombinase and yellow fluorescent protein (YFP) from adjacent, back-to-back Thy1 promoters. This tandem arrangement circumvents problems associated with trying to drive high expression of two genes from the same promoter. For the construct, first author Paul Young and colleagues used the CreER variant, which keeps Cre inactive in the absence of the estrogen receptor ligand, tamoxifen. This gives temporal control of recombinase activity (see image below).
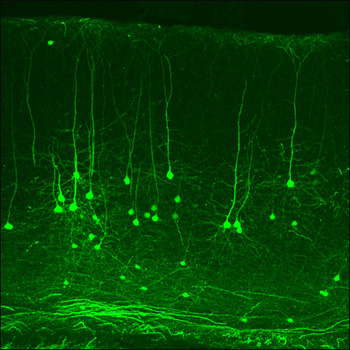
SLICK Neurons in Mouse Cortex
This slice of cortex from the SLICK-X mouse line shows that a subset of neurons express YFP (green) and the CreER recombinase. Treating these mice, or any offspring, with tamoxifen induces Cre activity and ablates any flox-flanked genes. Since Cre expression is limited to green neurons, the technique can be used to probe gene function in individual neurons. Image credit: Guoping Feng
The researchers validated the SLICK method using a Cre reporter mouse strain that expresses β-galactosidase on excision of an upstream stop codon by the recombinase (the R26R line). Crossing R26R with the SLICK mice produced offspring that expressed β-galactosidase on administration of tamoxifen, showing that the concept worked. To test the versatility of the method, Young and colleagues crossed SLICK-A mice, which show high Cre-YFP expression in motor neurons, with mice carrying a “floxed” conditional knockout allele for the choline acetyltransferase (ChAT) gene.
ChAT was undetectable in the YFP neurons four or eight weeks after administering tamoxifen, but interestingly, despite loss of ChAT, the neuromuscular junctions (NMJs) opposite the YFP nerve terminals remained intact and stained just as brightly for acetylcholine receptors as junctions from control animals. “This was a surprise, in the sense that there was no activity at all for such a long time. It suggests that adhesion molecules and cell-cell interactions are very important for stability, at least at the neuromuscular junction,” said Feng. He added that it will be interesting to see what happens in the central nervous system when cell activity is similarly silenced.
Feng already has experiments in mind to test this. He plans to use a method for silencing neurons described last year by David Anderson and colleagues at Caltech (see Lerchner et al., 2007). This is based on the expression of ivermectin-sensitive chloride channels. Administering ivermectin opens these channels, resulting in a flood of chloride that effectively quenches electrical activity. Expression of the chloride channel can be regulated by the Cre/lox system. A second option Feng is considering is based on G protein-coupled receptors developed by Brian Roth and colleagues at Case Western Reserve University, Cleveland, Ohio (see Armbruster et al., 2007). The G proteins are only activated by an inert drug-like compound. “Because these receptors are only activated by the drug, not endogenous ligands, if you express them in neurons you can either activate or inactivate the cells depending on which receptor you use,” said Feng.
Feng emphasized how easily the SLICK system can be used to test the contribution of virtually any gene to neuronal stability. Because many mouse genes are now, or soon will be, “floxed,” any of them could be crossed with the SLICK mice to probe gene function, suggested Feng. In this way, the role of neurotrophins, cytoskeleton proteins, or any number of genes could be tested. “Remember, not only can you do imaging, but also recording, because when you see the green neuron you know it is a knockout,” said Feng. He said that the SLICK mice will be a valuable tool for studying aging.
SLICK has the same goal as “mosaic analysis with double marker” (MADM), a method pioneered by Liqun Luo and colleagues at Stanford University (see Zong et al., 2005). Both genetically manipulate and label cells at the same time, but whereas SLICK can be used in mature animals, MADM depends on mitosis. This means that for studying neurons, MADM is most applicable to early developmental stages. MADM can, however, be used for non-neuronal cells and for all types of mutations, whereas SLICK is restricted to floxed conditional-knockout alleles. “Thus, although the goals of both methods are broadly similar, in practice they are likely to find different, but very complementary, applications in neuroscience research,” write Young and colleagues.
Feng’s group has reared 30 lines of SLICK mice, keeping mostly those that have a small subset of neurons expressing high levels of YFP. One of the lines, SLICK-V, shows brightly labeled individual neurons in the hippocampus and some sparse labeling of pyramidal neurons in the neocortex. Complete dendritic trees of CA1 hippocampal neurons are visible. Some of the lines are currently available at the Jackson Labs, and details are also on the Feng Lab website.
The work from Tsien and colleagues tackles a property that is often neglected when developing fluorescent probes—resistance to photobleaching. Though quantum yield and peak emission wavelength are often top of the wish list for new fluorescent proteins, all organic fluorophores wither under prolonged exposure to light. In daily lab practice, photobleaching limits repetitive imaging experiments.
To generate new photostable variants, first author Nathan Shaner and colleagues subjected genes for the fast-bleaching orange fluorescent protein mOrange to several rounds of random mutagenesis. They also tried a rational-design approach, using site-directed mutagenesis to change amino acids that interact with the chromophore on the red fluorescent protein, TagRFP. Photostability was screened by putting bacteria expressing the mutants under the spotlight—literally. The researchers shone a collimated beam of intense light on bacterial colonies for 10-20 minutes (colonies were kept cool), then measured their fluorescence.
The directed approach yielded TagRFP-T, which is ninefold more photostable than the parent, TagRFP, though only slightly less bright. The best fluorophore from the random mutagenesis screen was mOrange2. While it is about 30 percent less bright than the parent, mOrange2 is 25-fold more stable and twice as stable as the previous most photostable orange probe, KO. “We expect our screening method to be applicable to any of the existing fluorescent proteins, and, with modifications, to be useful in selecting for more efficient photoconvertible and photoswitchable fluorescent proteins as well,” write the authors. TagRFP-T may already be destined for the bright lights since it is the most photostable monomeric fluorescent protein of any color yet.—Tom Fagan
References
Paper Citations
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005 May 6;121(3):479-92. PubMed.
External Citations
Further Reading
No Available Further Reading
Primary Papers
- Young P, Qiu L, Wang D, Zhao S, Gross J, Feng G. Single-neuron labeling with inducible Cre-mediated knockout in transgenic mice. Nat Neurosci. 2008 Jun;11(6):721-8. PubMed.
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008 Jun;5(6):545-51. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.