Single-Cell Sleuthing Nabs Neurons Prone to Perish in Parkinson’s
Quick Links
The death of dopaminergic neurons in the substantia nigra is the defining feature of Parkinson’s disease, but not all neurons in the region suffer this fate. Why do some wither while others survive until the bitter end? A study published May 5 in Nature Neuroscience opens the door to answers. Working from a cutting-edge transcriptomic toolbox, researchers led by Evan Macosko at the Broad Institute of Harvard and MIT in Cambridge, Massachusetts, defined 10 transcriptionally distinct populations of dopaminergic neurons in the human substantia nigra pars compacta. One—marked by its expression of SOX6 and AGTR1—selectively succumbed to degeneration in people with PD and dementia with Lewy bodies (DLB). These cells reside in the ventral part of the SN, a region known for its susceptibility to neurodegeneration. What’s more, the cells expressed more PD risk genes than did any other dopaminergic neuron subtype, suggesting that neurodegeneration in these synucleinopathies is a cell-intrinsic process, as opposed to one inflicted by other cell types.
- Single-nucleus profiling uncovered 10 transcriptional subtypes of dopaminergic neuron in the human substantia nigra pars compacta.
- One, expressing SOX6 and AGTR1, was hit hardest in people with PD or DLB.
- This vulnerable subset expressed more PD risk genes than any other subtype of dopaminergic neuron.
“It is a great accomplishment to reach the level of cell-specific localization of the genes most clearly associated with PD,” wrote Ann Graybiel of the Massachusetts Institute of Technology. “This article opens a new window for work on the root causes of Parkinson’s disease, and a glimpse of how this may differ from Alzheimer’s disease.”
That some dopaminergic (DA) neurons survive while others die over the course of Parkinson’s disease has been known for decades, yet pinning down the molecular mechanisms underlying this selective vulnerability is challenging (Yamada et al., 1990; Gibb and Lees, 1991; Lu et al., 2006; May 2009 news). Single-cell RNA-Sequencing techniques enable scientists to define the gene-expression profiles that distinguish hardy from fragile neurons. However, the diminutive size of the substantia nigra pars compacta, coupled with the throngs of cell types within and surrounding it, make teasing out DA neurons, let alone different subsets of them, a finicky endeavor. Previous single-cell transcriptomic studies have had to make do with vanishingly small numbers of dopaminergic neurons, making it hard to detect distinct subsets in the first place, much less robustly compare them across samples (Agarwal et al., 2020; Smajić et al., 2022).
To harvest many more nuclei from postmortem brain samples, co-first authors Tushar Kamath and Abdulraouf Abdulraouf and colleagues first fished for unique nuclear markers they could use to enrich for these nuclei. They ultimately pegged Nr4a2, a transcription factor only DA neurons express. Then they isolated nuclei from postmortem substantia nigra samples of eight people who had died without neurodegenerative disease. Selecting with Nr4a2, the researchers netted 5,684 DA neuron nuclei for RNA-Sequencing analysis—topping the haul from previous studies by 180-fold.

Fishing for DA Nuclei. Scientists carved out the substantia nigra pars compacta from postmortem brain, isolated the nuclei, and used the NR4A2 protein to snag those that came from DA neurons for transcriptomic profiling. [Courtesy of Kamath et al., Nature Neuroscience, 2022.]
Using LIGER, a computational method developed in the lab to compare transcriptomes from multiple samples, Kamath pulled 10 distinct transcriptional subsets from among the DA neuron herd. These subsets varied in their expression of two known DA developmental markers, SOX6 and CALB1, such that four subsets preferentially expressed SOX6, while six leaned toward CALB1.
The researchers identified 10 similar transcriptomic subsets of DA neurons within the macaque substantia nigra. Intriguingly, in mice and rats they detected all but one of these subsets—marked by its expression of CALB1 and GEM—suggesting that it occurs only in primates. The scientists are studying this subset further, on the hunch that it could be behind a nigral circuit with the cortex that has been seen in primates but not rodents. Macosko’s team is studying what this circuit might be doing, and whether it relates to neurodegenerative disease.
Exactly where within the substantia nigra were these different types of DA neuron? To find out, the researchers applied a technique called Slide-Seq to sections of the macaque substantia nigra, which is less than half as big the human SN but arranged similarly. The method works like this: Scientists first create an array of beads, each coated with a unique DNA sequence, or “barcode,” that enciphers the bead’s spatial location within the array. Then, the scientists apply a frozen slice of brain tissue to the array, so that RNA from within the tissue transfers onto the beads. By sequencing the RNA as well as the DNA barcode attached to each bead, researchers can discern the address of individual transcriptomes within the tissue section. In this way, the researchers were able to locate all 10 subsets of DA neurons within the macaque SNpc. Notably, one subset, marked by its expression of both SOX6 and AGTR1, resided entirely within the ventral segment of the substantia nigra, an area known to degenerate in PD.

Find My Subtype. RNA from regions (circle) of the macaque substantia nigra pars compacta (left) were transferred to bead arrays DNA-barcoded for spatial transcriptomics. Both ventral and dorsal halves of the SNpc contain DA neurons (top, black), but their subtypes were distributed differently across the SNpc (middle and bottom panels). The light blue ones are most prone to die in PD. [Courtesy of Kamath et al., Nature Neuroscience, 2022.]
To pinpoint which of these subsets might be most prone to perish in neurodegenerative disease, the researchers performed snRNA-Seq on substantia nigra samples from 10 people who had died with PD or DLB. Unsurprisingly, people with PD or DLB had a dearth of DA neurons relative to healthy donors. The AGTR1 subset was the most depleted, suggesting that this type of DA neuron is particularly vulnerable to neurodegeneration. In contrast, two other DA neuron subsets, both expressing CALB1, were relatively more preserved in people with PD/DLB.
A subset of microglia expressing GPNMB, a known marker of disease-associated microglia associated with AD, was expanded, as was a subset of astrocytes. The findings reflect overlapping neurodegenerative mechanisms in PD and DLB, which are both synucleinopathies but have important differences, as well. As Ernest Arenas of the Karolinska Institute in Stockholm noted in a commentary that accompanied the paper, people with DLB have cognitive symptoms, along with Aβ plaques and tau tangles, in addition to dopaminergic neuron loss and its attendant motor symptoms. “In this context, the analysis provides valuable information on common alterations in these two diseases, but some disease-specific alterations may be underrepresented and undetected. Future analysis of more samples from individuals with PD or DLB will help to clarify this point,” Arenas wrote.
To address the question of what made the AGTR1 subset of DA neurons so susceptible to the ravages of PD or DLB, the scientists searched the transcriptomes of nigral cell types for their expression of genes tied to PD risk. These included 26 genes for which rare familial PD variants had been identified, as well as myriad common variants that had been tied to PD risk in genome-wide association studies.
They found that, as a whole, DA neurons expressed more of these PD risk genes than did any other cell type. Among the DA neurons, the largest and only statistically significant enrichment of PD genetic risk genes was in the AGTR1 subtype.
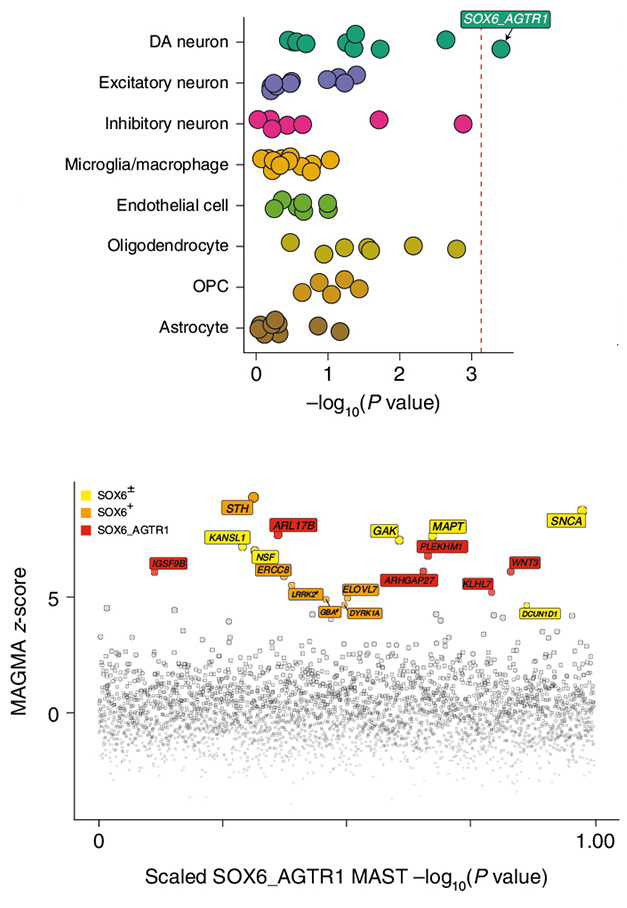
Doomed to Degenerate? Transcriptional subsets of different cell types expressed different levels of genes tied to PD. Of the 10 dopaminergic subsets, only the AGTR1+ subset was significantly enriched for PD risk (top graph). Different PD risk genes were enriched in SOX6+AGTR1+ cells (red), SOX6+ cells (orange), or dopaminergic neurons more broadly (yellow). [Courtesy of Kamath et al., 2022.]
What are the cellular pathways active in the AGTR1+ DA neuron subset that might seal their fate in PD? This remains unclear, but the scientists have some suspects, including genes involved in cell stress and mitochondrial dysfunction that were ramped up in this vulnerable subtype relative to others. AGTR1 itself encodes the angiotensin receptor 1 and may play a role, Arenas noted in his commentary. Variants in the gene have been tied to PD risk in a Japanese population, and inhibitors of the renin-angiotensin system are known to boost motor function in people with PD (Saito et al., 2003; Reardon et al., 2000; Labandeira-García et al., 2014).
In contrast to dopaminergic neurons, glial cells did not express an overabundance of PD risk genes. Instead, these cells tended to express genes tied to AD risk. The authors interpreted these findings as evidence that neurodegeneration in PD is primarily a cell-intrinsic process, while in AD, neuronal death is more tied to the actions of other cell types including microglia.
To Macosko, this is the most important insight from the study. “The specific enrichment of genetic heritability is in the cells that die,” he said. “This suggests the disease is somewhat cell autonomous.”
Ole Isacson of Harvard Medical School in Boston took a different view. Noting that most PD is sporadic, meaning it arises from external stressors interacting with vulnerable dopaminergic neurons, he interprets the finding that mitochondrial and cell stress pathways were revved in this vulnerable subset as a sign that the cells are under duress. Because dopaminergic neurons of the SNpc are known to broil with quadruple the level of oxidative stress as their counterparts in nearby regions, the effects of that stress over a lifetime could build up to render the cells vulnerable to external insults such as environmental toxins and neuroinflammation, he believes. Isacson sees the proportional increase in GPNMB-expressing microglia as a sign of an ongoing inflammatory response to lipid stress in the SNpc (Brekk et al., 2020). His data, combined with studies indicating that TNF-α inhibitors reduce PD risk, implicates parallel processes of inherent stress within vulnerable dopaminergic neurons and rising neuroinflammatory insults that may reach a neurodegenerative "boiling point" with age (Hallett et al., 2019).
Mark Cookson at the National Institutes of Health in Bethesda, Maryland, agrees that the PD risk gene analysis needs further investigation, noting that several genes considered in the analysis have uncertain ties to PD. He was surprised to see such strong expression of LRRK2 in the vulnerable neurons, given that other single-cell transcriptomic studies have found this PD gene to be strongly expressed in microglia and oligodendrocyte precursor cells (e.g., Wang et al., 2022). “It would therefore be important to concatenate multiple nigral datasets … to try to understand which results are most consistent between sample series.”
“Preferential vulnerability is one of the major mysteries of neurodegenerative diseases,” commented Clemens Scherzer of Brigham and Women’s Hospital in Boston “This exciting study takes a hard look at identifying preferentially vulnerable dopamine neuron subtypes in the midbrain and provides intriguing new insights on possible subtypes of pars compacta dopamine neurons.”
However, Scherzer noted that while the study addresses differences among dopaminergic neurons within the SNpc, it does not explain why SNpc DA neurons are more vulnerable to PD than their counterparts in the neighboring ventral tegmental area. “The most resistant dopamine neurons in the midbrain, the VTA dopamine neurons, do not seem to have been included [in the study]. Does this kick the can down the road?” Scherzer asked (full comment below).
Macosko told Alzforum that the lab is currently investigating the transcriptomes of dopaminergic neurons located in different regions of the brain, and also in the peripheral nervous system. Some scientists believe that α-synuclein aggregation in PD starts outside of the brain, within neurons of the gut, where it hitches a ride into the CNS via the vagus nerve (Jul 2011 news). Comparing the gene-expression profiles of neurons posted along this proposed route might give researchers clues about the likelihood of this disease progression model, Macosko suggested.
He also hopes scientists will plumb the current dataset for therapeutic targets, design more accurate disease models, and even to differentiate the most needed types of dopaminergic neurons for cell replacement therapies.—Jessica Shugart
References
News Citations
- Multiple Hits Explain Selective Loss of Dopamine Neurons in PD
- Parkinson's: It Started With a Gut Feeling
Paper Citations
- Yamada T, McGeer PL, Baimbridge KG, McGeer EG. Relative sparing in Parkinson's disease of substantia nigra dopamine neurons containing calbindin-D28K. Brain Res. 1990 Sep 3;526(2):303-7. PubMed.
- Gibb WR, Lees AJ. Anatomy, pigmentation, ventral and dorsal subpopulations of the substantia nigra, and differential cell death in Parkinson's disease. J Neurol Neurosurg Psychiatry. 1991 May;54(5):388-96. PubMed.
- Lu L, Neff F, Fischer DA, Henze C, Hirsch EC, Oertel WH, Schlegel J, Hartmann A. Regional vulnerability of mesencephalic dopaminergic neurons prone to degenerate in Parkinson's disease: a post-mortem study in human control subjects. Neurobiol Dis. 2006 Aug;23(2):409-21. Epub 2006 Jun 6 PubMed.
- Agarwal D, Sandor C, Volpato V, Caffrey TM, Monzón-Sandoval J, Bowden R, Alegre-Abarrategui J, Wade-Martins R, Webber C. A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat Commun. 2020 Aug 21;11(1):4183. PubMed.
- Smajić S, Prada-Medina CA, Landoulsi Z, Ghelfi J, Delcambre S, Dietrich C, Jarazo J, Henck J, Balachandran S, Pachchek S, Morris CM, Antony P, Timmermann B, Sauer S, Pereira SL, Schwamborn JC, May P, Grünewald A, Spielmann M. Single-cell sequencing of human midbrain reveals glial activation and a Parkinson-specific neuronal state. Brain. 2022 Apr 29;145(3):964-978. PubMed.
- Saito S, Iida A, Sekine A, Kawauchi S, Higuchi S, Ogawa C, Nakamura Y. Catalog of 178 variations in the Japanese population among eight human genes encoding G protein-coupled receptors (GPCRs). J Hum Genet. 2003;48(9):461-468. Epub 2003 Aug 30 PubMed.
- Reardon KA, Mendelsohn FA, Chai SY, Horne MK. The angiotensin converting enzyme (ACE) inhibitor, perindopril, modifies the clinical features of Parkinson's disease. Aust N Z J Med. 2000 Feb;30(1):48-53. PubMed.
- Labandeira-García JL, Garrido-Gil P, Rodriguez-Pallares J, Valenzuela R, Borrajo A, Rodríguez-Perez AI. Brain renin-angiotensin system and dopaminergic cell vulnerability. Front Neuroanat. 2014;8:67. Epub 2014 Jul 8 PubMed.
- Brekk OR, Honey JR, Lee S, Hallett PJ, Isacson O. Cell type-specific lipid storage changes in Parkinson's disease patient brains are recapitulated by experimental glycolipid disturbance. Proc Natl Acad Sci U S A. 2020 Nov 3;117(44):27646-27654. Epub 2020 Oct 15 PubMed.
- Hallett PJ, Engelender S, Isacson O. Lipid and immune abnormalities causing age-dependent neurodegeneration and Parkinson's disease. J Neuroinflammation. 2019 Jul 22;16(1):153. PubMed.
- Wang Q, Wang M, Choi I, Ho L, Farrell K, Beaumont KG, Sebra R, Crary JF, Davis DA, Sun X, Zhang B, Yue Z. Single-cell transcriptomic atlas of the human substantia nigra in Parkinson’s disease. bioRxiv. March 30, 2022 bioRxiv
Further Reading
Primary Papers
- Kamath T, Abdulraouf A, Burris SJ, Langlieb J, Gazestani V, Nadaf NM, Balderrama K, Vanderburg C, Macosko EZ. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson's disease. Nat Neurosci. 2022 May;25(5):588-595. Epub 2022 May 5 PubMed.
- Arenas E. Parkinson's disease in the single-cell era. Nat Neurosci. 2022 May;25(5):536-538. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
Massachusetts Institute of Technology
This article opens a new window for work on the root causes of Parkinson’s disease and a glimpse of how this may differ from Alzheimer’s disease. The authors used advanced snRNA-sequencing and related methods on a special population of PD brain samples and normative control samples. They identified the transcriptomic identities of the dopamine-containing substantia nigra neurons most vulnerable in PD. These express particular patterns of transcription factors, and can be identified for the most part across species, with mice lacking some of the markers conserved across macaques and humans.
Crucially, the authors used spatial-seq to enable them to view the transcriptionally vulnerable neurons in anatomical maps of the substantial nigra. Because of this multimodal approach, they were able to achieve the so-far-rare goal of relating their transcriptional identification of the vulnerable neurons to the actual locations of the vulnerable neurons in the brain locus examined (here, substantial nigra). The remarkable result is that they have identified as the most transcriptionally vulnerable cell group the very most ventral zone originally earmarked as most vulnerable by classical neurochemical anatomical studies in postmortem PD patients, and scored along with other nigral zones in terms of a sequential vulnerability in the progression of PD.
The most vulnerable region, in the “ventral tier” of the substantial nigra, is identifiable by expression of Aldh1A1, which is a clear marker of the ventral tier from rodent to humans. The tanscriptomic identifier is AGTR1, alongside the nigrosome marker pattern of low Calbindin. The authors give strong evidence that these features are the crucial ones, making it likely that they actually are true identifiers of a cell type—what is called an endogenous or cell-autonomous feature.
These are crucial facts that offer great potential for understanding and treating PD. This is because the ventral tier dopamine-containing neurons of the substantial nigra receive preferential input from the striosomal compartment of the striatum. This compartment has been linked to mood-related problems that frequently occur early on in PD, and it has molecularly specialized dopaminergic characteristics itself.
The early and strong vulnerability of this set of nigral dopamine-containing neurons could be a key to understanding the molecular and phenotypic expression of PD. It now will be possible to build a systems-neuroscience strategy to combat and defeat PD, and it will be possible then to design therapies including the use of enhancers and novel transport methods for therapeutic delivery.
It is a great accomplishment to reach the level of cell-specific localization of the genes most clearly associated with PD. The authors' comparison with the case of AD is also illuminating—here, there is a local reference, compared with more global involvement in the case of advanced Alzheimer’s disease.
National Institute on Aging
This paper is an impressive application of two types of single-cell analysis, especially when considering the additional flow sorting step to enrich for dopamine neurons in figure 1. That the general dorsal/ventral gradient of calbindin vs. SOX6 is replicated is reassuring, and the identification of susceptible subtypes that are marked by SOX6 and AGTR1 adds another dimension to what we know about DA neurons sensitivity.
One area that I think needs some discussion is the genetic enrichment analyses. It is consistent with other recent enrichment analyses that suggest dopamine neurons account for some of the genetic risk of PD (Bressan et al., 2021). That said, some of the presented genes for PD in Kamath et al.’s analysis are uncertain, e.g. UCHL1, GUGYF2, HTRA2, EIF4G1, etc., hence how well the enrichment scores would work on a smaller gene set would be important to understand.
Additionally, it was very surprising to see such a strong effect of LRRK2, especially in contrast to other recent surveys that indicated stronger expression in microglia and oligodendrocyte precursor cells in human substantia nigra (Wang et al., 2022). It would therefore be important to concatenate multiple nigral datasets using different single nuclear/cell RNA-Seq approaches to try to understand which results are most consistent between sample series.
References:
Bressan E, Reed X, Bansal V, Hutchins E, Cobb MM, Webb MG, Alsop E, Grenn FP, Illarionova A, Savytska S, Violich I, Broeer S, Fernandes N, Sivakumar R, Beilina A, Billingsley K, Berghausen J, Pantazis CB, Meechoovet B, Reiman R, Courtright-Lim A, Logemann A, Antone J, Barch M, Kitchen R, Li Y, Dalgard CL, The American Genome Center, Rizzu PR, Hernandez DG, Hjelm BE, Nalls M, Gibbs JR, Finkbeiner S, Cookson MR, Van Keuren-Jensen K, Craig DW, Singleton AB, Heutink P, Blauwendraat C. The Foundational data initiative for Parkinson’s disease (FOUNDIN-PD): enabling efficient translation from genetic maps to mechanism. BioRxiv, June 3, 2021 bioRxiv
Wang Q, Wang M, Choi I, Ho L, Farrell K, Beaumont KG, Sebra R, Crary JF, Davis DA, Sun X, Zhang B, Yue Z. Single-cell transcriptomic atlas of the human substantia nigra in Parkinson’s disease. bioRxiv. March 30, 2022 bioRxiv
Harvard Medical School, Brigham and Women's Hospital
Preferential vulnerability is one of the major mysteries of neurodegenerative diseases. This exciting study takes a hard look at identifying preferentially vulnerable dopamine neuron subtypes in the midbrain and provides intriguing new insights on possible subtypes of pars compacta dopamine neurons.
The study raises several important questions:
The substantia nigra pars compacta dopamine (SNpc) neurons analyzed are all vulnerable to PD, although to different degrees. The most resistant dopamine neurons in the midbrain, the VTA dopamine neurons, do not seem to have been included. Does this kick the can down the road? What makes SNpc dopamine neurons more vulnerable than VTA dopamine neurons?
Do the 10 nominated SNpc dopamine neuron subtypes truly represent replicable, discrete subtypes rather than various presentations of the population spectrum of pars compacta neurons? The putative subtype clusters appear closely linked and interwoven rather than clearly separated. Considering the evolving “art and science of clustering,” this view might evolve as we learn more about their validity and the influence of covariates in larger cohorts.
Generally, much larger sample sizes will help to solidify these intriguing clues. Caveats are the limited number of eight control midbrains, six of which were female, and the huge age range they represent, which did not seem to match the PD cases very well.
This is an exciting time for brain research. Single-nucleus transcriptomics opens the awesome opportunity to map the circuit board of Parkinson’s with single-cell and single base-pair resolution. The PD5D Consortium at Harvard, the Broad Institute, and the University of Wisconsin—part of the ASAP Collaborative Research Network—have launched the ambitious initiative to map a Parkinson Cell Atlas in five dimensions based on spatial and single-nucleus multiomics of a million brain cells with the ultimate goal to map, simulate, and help to correct the flow of genetic information from patient genomes to brain cells in brain space.
Harvard Medical School
Harvard Medical School
Neighboring but functionally distinct dopaminergic neurons are vulnerable or resistant to neurodegeneration during Parkinson’s disease. Over the last 20 years, we and others have used cell-type-specific analyses to characterize healthy neurons and to propose causal pathways of degeneration.
These pathways include intrinsic oxidative stress, where, for example, live-cell imaging revealed that intraneuronal oxidation levels are greater in vulnerable dopaminergic neurons than in disease-resistant dopaminergic neurons (Guzman et al., 2010). Vesicular trafficking and mitochondrial pathways were highlighted when RNA was collected from populations of vulnerable and disease-resistant dopaminergic neurons (Chung et al., 2005). Image-based analyses demonstrated several converging pathways as healthy ventral-tier dopaminergic neurons age that could be consistent with changes in mitochondrial DNA over time (Chung et al., 2005; Dölle et al., 2016; Collier et al., 2011).
Beyond dopaminergic neuron subtypes, image-based analysis of human postmortem tissue revealed lipid accumulation in nigral dopaminergic neurons and microglia but not astrocytes (Brekk et al., 2020). In the same study, we found correlations between triglyceride and GPNMB levels in nigral tissue (Brekk et al., 2020).
During this period, we avoided molecular analyses of degenerating neurons because we felt that causality could not be measured in individual cells adapting in an asynchronous, protracted manner to a disease driver, ultimately leading to a collective loss of circuit function once the threshold of synaptic loss has been passed (Figure 1) (Engelender et al., 2017).
In this new work, Tushar Kamath, Abdulraouf Abdulraouf and colleagues provide an excellent open-access catalog of transcripts from different types of aged dopaminergic neurons isolated from the brains of individuals without neurological symptoms (Kamath et al., 2022). The single-cell data is broadly consistent with previous studies and methods (Mendez et al., 2005; Kadkhodaei et al., 2009; Thompson et al., 2005), while providing greater resolution. The data instantly fills gaps for drug discovery. For example, the initial steps of drug and biomarker discovery can be helped if a molecular target is known to be expressed by the target cell population.
However, we continue to believe that causality cannot be interpreted from the transcriptome of degenerating patient neurons. The expression of genes associated with PD risk by ventral tier dopaminergic neurons is an important step when considering how to use “GWAS hits,” but we expect these genes to be expressed in different combinations across many different cell types of the body and under different dysregulated states. This concept needs experimental determination, but hints at underlying physiological processes in vulnerable neurons that drive expression of these genes under periods of neuronal challenge (“biomarkers of a dysregulated state”).
Kamath et al. also propose to use the single-cell transcriptional data to improve in-vitro differentiation protocols for disease modeling. This idea seems like a bit of a stretch given that the cellular environment and functional state of the neurons in the aged brain are so strikingly different than cell culture, be it as a two- or three-dimensional culture. Even cells cultured over a couple of years are unlikely to reach toward the aged end of the structure and function spectrum that would be reflected in their transcriptome.
Figure 1. Transcriptional profiles within a population of aged, vulnerable neurons undergoing asynchronous neurodegeneration. (A,B) Melanized neurons of the young (A) or aged (B) ventral midbrain are a population of dopaminergic neurons that are vulnerable to neurodegeneration in Parkinson’s disease (PD). (C) Fewer melanized neurons are a characteristic of the ventral midbrain from a PD patient. (D) Hypothetical transcriptional diversity of the vulnerable neurons in the young healthy brain shows a largely homeostatic profile (green; most cell-cell transcriptional variability is caused by asynchronous, physiological responses to functional challenges). (E) Neurons begin to show dysregulated transcriptional profiles during aging that represent a significant and potentially lethal challenge to an individual cell but too few cells are lost to compromise the functional threshold of synaptic loss and cause symptoms unless the challenge persists over a protracted period (yellow-red). (F) The hypothetical transcriptional diversity in the PD patient population of degenerating neurons is again asynchronous and shows greater cell-cell variation and dysregulation as the cells reroute transcriptional activity to adapt and survive the disease driver.
References:
Guzman JN, Sanchez-Padilla J, Wokosin D, Kondapalli J, Ilijic E, Schumacker PT, Surmeier DJ. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010 Dec 2;468(7324):696-700. PubMed.
Chung CY, Seo H, Sonntag KC, Brooks A, Lin L, Isacson O. Cell type-specific gene expression of midbrain dopaminergic neurons reveals molecules involved in their vulnerability and protection. Hum Mol Genet. 2005 Jul 1;14(13):1709-25. PubMed.
Dölle C, Flønes I, Nido GS, Miletic H, Osuagwu N, Kristoffersen S, Lilleng PK, Larsen JP, Tysnes OB, Haugarvoll K, Bindoff LA, Tzoulis C. Defective mitochondrial DNA homeostasis in the substantia nigra in Parkinson disease. Nat Commun. 2016 Nov 22;7:13548. PubMed.
Collier TJ, Kanaan NM, Kordower JH. Ageing as a primary risk factor for Parkinson's disease: evidence from studies of non-human primates. Nat Rev Neurosci. 2011 Jun;12(6):359-66. PubMed.
Brekk OR, Honey JR, Lee S, Hallett PJ, Isacson O. Cell type-specific lipid storage changes in Parkinson's disease patient brains are recapitulated by experimental glycolipid disturbance. Proc Natl Acad Sci U S A. 2020 Nov 3;117(44):27646-27654. Epub 2020 Oct 15 PubMed.
Engelender S, Isacson O. The Threshold Theory for Parkinson's Disease. Trends Neurosci. 2017 Jan;40(1):4-14. Epub 2016 Nov 25 PubMed.
Kamath T, Abdulraouf A, Burris SJ, Langlieb J, Gazestani V, Nadaf NM, Balderrama K, Vanderburg C, Macosko EZ. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson's disease. Nat Neurosci. 2022 May;25(5):588-595. Epub 2022 May 5 PubMed.
Mendez I, Sanchez-Pernaute R, Cooper O, Viñuela A, Ferrari D, Björklund L, Dagher A, Isacson O. Cell type analysis of functional fetal dopamine cell suspension transplants in the striatum and substantia nigra of patients with Parkinson's disease. Brain. 2005 Jul;128(Pt 7):1498-510. PubMed.
Kadkhodaei B, Ito T, Joodmardi E, Mattsson B, Rouillard C, Carta M, Muramatsu S, Sumi-Ichinose C, Nomura T, Metzger D, Chambon P, Lindqvist E, Larsson NG, Olson L, Björklund A, Ichinose H, Perlmann T. Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J Neurosci. 2009 Dec 16;29(50):15923-32. PubMed.
Thompson L, Barraud P, Andersson E, Kirik D, Björklund A. Identification of dopaminergic neurons of nigral and ventral tegmental area subtypes in grafts of fetal ventral mesencephalon based on cell morphology, protein expression, and efferent projections. J Neurosci. 2005 Jul 6;25(27):6467-77. PubMed.
Make a Comment
To make a comment you must login or register.