Noncoding RNAs Evince World of Gene Regulation in Dopaminergic Neurons
Quick Links
Emanating from the largely unknown vastness of the non-protein-coding genome are tens of thousands of unique transcripts that point to a hidden world of gene regulation in human neurons. Researchers led by Clemens Scherzer of Brigham and Women’s Hospital in Boston deployed both laser-capture technology and RNA sequencing to detect an abundance of transcribed noncoding elements (TNEs) in specific subsets of neurons in the human brain. Read largely from intronic DNA, many of these elements hailed from regions of the genome bearing markers of active enhancers—sequences that boost the expression of other genes. While it is unclear if TNEs have a function in their own right, at the least they are useful proxies for measuring the activity of enhancers, claim the authors. Their paper, published September 17 in Nature Neuroscience, reports that variants identified in genome-wide association studies (GWAS) of dopaminergic diseases, including Parkinson’s, alter the expression of TNEs. In particular, PD risk variants at the 17q21 locus appeared to dial up expression of one TNE in the KANSL1 gene.
- Dopaminergic neurons transcribe 70,000+ noncoding elements.
- Most are transcribed from introns, a third from active enhancers.
- Genetic variants tied to diseases change their expression.
“This result is exciting because it ascribes a functional role to these GWAS hits for the first time. It opens the door for future discovery and characterization of which genes these enhancers regulate, how they work at the mechanistic level, and how they are dysregulated in disease states,” commented Jeremy Day of the University of Alabama in Birmingham.
“Classifying gene-expression patterns and their relationship to genetic variants is a very important way to determine proximate genetic mechanisms arising from GWAS,” commented Mark Cookson of the National Institutes of Health in Bethesda. “The use of laser capture microdissection and RNA-Seq is an interesting way to generate these types of datasets.”
The vast majority of variants detected in GWAS fall within regions that encode no protein, implying that gene regulation drives most disease risk. Scientists try to understand this by examining how risk variants correlate with expression of nearby genes, or with markers of protein-based enhancer activity such as histone acetylation (e.g., Apr 2018 news; Jul 2018 news). But the problem is even more complicated. Researchers have discovered that enhancer regions not only regulate transcription of downstream genes, but can be transcribed into RNA in their own right. Apparently, when enhancers recruit the transcriptional machinery to activate a gene, they, too, can be transcribed. (Kim et al., 2010; Andersson et al., 2014). Hence, these transcripts can serve as proxies for enhancer activity. However, enhancer RNAs are commonly overlooked in RNA sequencing studies. Researchers have yet to catalogue them and examine how they relate to disease variants in different cell types.
First author Xianjun Dong and colleagues sought to do just that. Given that dopaminergic neurons are vulnerable in PD and other diseases, the researchers chose to characterize RNA species in these cells and see if their expression correlated with disease-associated genetic variants. Dong and second author Zhixiang Liao embarked on a titanic task, said Scherzer. Using laser capture microdissection, they plucked 40,000 dopaminergic neurons from the substantia nigra of 86 postmortem brains. For comparison, they also dissected out pyramidal neurons from the temporal cortices of 10 brains and from the motor cortices of three brains, and collected fibroblast and white blood cell samples as well. Dong and Liao then sequenced the pooled RNA from each cell type, including mRNAs and all noncoding sequences hailing from introns and intergenic regions. All of the brain samples came from people who had died free of a neuropsychiatric or neurodegenerative disease.
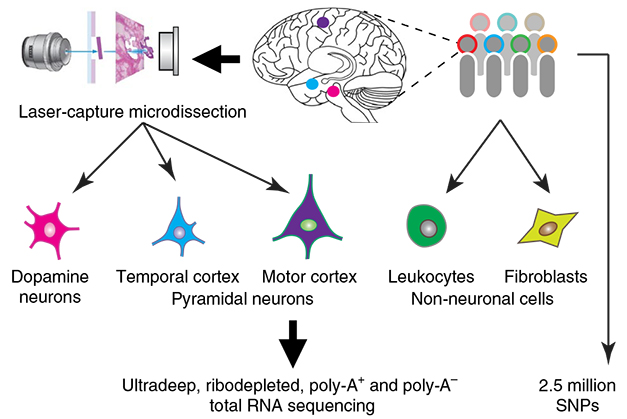
Laser capture RNA-Seq. In lcRNASeq, researchers use a laser to dissect out individual neurons and other cells, then sequence their total RNA. [Courtesy of Dong et al., Nature Neuroscience, 2018.]
The researchers found that 64 percent of the entire human genome was transcribed in some form or another in these dopaminergic neurons. Scherzer called this number “mind-boggling.” More than half of these transcripts derived from intergenic regions and introns.
How many were transcribed enhancers? Instead of limiting their search to previously established enhancer sequences, the researchers first selected the TNE class of transcripts. These transcripts arose from regions other than exons or promoters, were at least 100 base pairs in length, and were expressed broadly across brain samples. Nearly a third of all transcripts from dopaminergic and pyramidal neurons qualified as TNEs, compared with a fifth of transcripts from the non-neuronal cells.

TNE Multiverse.
Cell types expressed a unique signature of transcribed noncoding elements as well as common TNEs. [Courtesy of Dong et al., Nature Neuroscience, 2018.]
In dopaminergic neurons, the researchers detected more than 71,000 TNEs, compared with 37,000 in pyramidal neurons, and 19,000 in non-neuronal cells. Perhaps surprisingly, most of the TNEs—92 percent—resided in introns. A third of the TNEs in dopaminergic neurons were transcribed from DNA stretches previously reported to bear at least one hallmark of an active enhancer—such as histone modification, DNA methylation, or chromatin accessibility. This was true in the human cortical samples and in neuroblastoma cells, which are sometimes used as cellular models for PD.
It is unclear what the other two-thirds of TNEs represent, but Scherzer speculated that they could be transcribed from enhancers that are uniquely active in dopaminergic neurons. Importantly, each cell type had a distinct TNE complement, and this could be used to distinguish between dopaminergic, pyramidal, and non-neuronal samples. Among 106 individual postmortem samples, the signature accurately designated the correct cell type 99.1 percent of the time. Messenger RNA and noncoding RNA signatures clustered cell types with a similar accuracy.
The researchers next asked whether any disease-associated variants identified in GWAS localized to genomic regions encoding TNEs or other RNA species. Sifting through variants linked to more than 1,000 diseases in the National Human Genome Research Institute GWAS catalog (Welter et al., 2014), they found 61 diseases enriched for variants that landed within regions encoding dopaminergic TNEs. By comparison, 43 diseases had an abundance of variants that hailed from other noncoding RNAs, and just 11 diseases had variants that resided in exons.
Among these 61 diseases, 11 were dopaminergic disorders or traits, including PD, schizophrenia, addiction, and bipolar disorder. Variants linked to diseases more loosely associated with the dopaminergic system, such as cardiovascular disease, obesity, diabetes, and sleep disorders, also landed in dopaminergic TNEs. TNEs also harbored many variants known to alter the effectiveness of dopaminergic drugs.

TNEs and Disease.
Genetic variants linked to a variety of disease were found more often in TNEs than in exons or promoters. [Courtesy of Dong et al., Nature Neuroscience, 2018.]
The researchers next asked whether genetic variation affected the expression of TNEs. An expression quantitative trait loci (eQTL) analysis of 84 postmortem brain samples matched genetic variants with expression of TNEs within a megabase. Among more than 4.2 million single-nucleotide polymorphisms analyzed, nearly 3,500 affected expression of 151 TNEs, which resided in the introns of 102 unique genes. Of these genes, a large proportion were involved in synaptic function. By comparison, 3,300 polymorphisms affected the expression of 52 noncoding RNAs, and 676 variants altered the expression of 46 mRNAs.
The findings suggested that genetic variants influence TNE expression. To test whether this holds true for disease-associated variants, the researchers correlated nearly 500,000 polymorphisms tied to more than 1,000 human diseases with levels of TNE expression—again just in dopaminergic neurons. They found that nearby disease variants altered expression of 23 TNEs.
Strikingly, six PD risk variants clustered on chromosome 17q21 correlated with increased expression of a TNE in the second intron of the KANSL1 gene. Variants at 17q21are strongly tied to PD, but it has been unclear exactly which gene confers that risk. Previous studies have implicated MAPT, the gene encoding tau; however, Scherzer’s analysis pulled up no association between the variants and expression of TNEs in or near the tau gene, nor of expression of the tau mRNA itself. The function of KANSL1 is unknown; deletions in the gene result in Koolen-de Vries syndrome, a neurological disease with severe learning disabilities.
Cookson was unconvinced that the data pin KANSL1 is the source of PD risk at the 17q21 locus. He notes data from the Genotype-Tissue Expression project suggest both KANSL1 and MAPT expression are affected by the PD risk polymorphisms (Oct 2017 news). “Therefore, depending on what evidence one might examine, one could favor either KANSL1 or MAPT,” he wrote, adding that more than one gene may confer risk from the locus.
Scherzer countered that GTEX and other body-wide transcriptomic studies such as FANTOM5 analyze RNA from different brain regions and do not specifically pick out dopaminergic neurons for analysis. They may have missed the regulation specific to dopamine neurons at this locus. GTEX relies on levels of mRNA expression as opposed to looking at TNEs, to gauge regulatory effects of variants, Scherzer added.
Cookson questioned whether dopaminergic neurons are the best place to look for regulatory changes that incite Parkinson’s. After all, immune cells such as microglia are involved in PD and other neurodegenerative diseases. “We should keep an open mind as to potential mechanisms that might include non-neuronal cells as well as multiple neuronal types, especially if the Braak hypothesis is correct and PD starts in the enteric or olfactory systems,” Cookson wrote. Scherzer acknowledged that gene-expression changes in other cell types could boost risk for PD, but thinks the unique susceptibility of dopaminergic neurons to the disease process likely plays a pivotal role.
The TNE data is publicly available. Scherzer hopes it will provide an entry point for researchers to investigate the complex epigenetics of dopaminergic disorders. The laser capture-TNE approach could yield similar insight into the regulatory mechanisms that underlie neurodegeneration more broadly. In future studies, Scherzer plans to include postmortem samples from people who had Parkinson’s and other diseases.—Jessica Shugart
References
News Citations
- GWAS, GWAX: bioRχiv Hosts Bonanza of Alzheimer’s Genetics
- A Delicate Frontier: Human Microglia Focus of Attention at Keystone
- Gene Expression Map of Human Body Gives Value to Variants
Paper Citations
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010 May 13;465(7295):182-7. Epub 2010 Apr 14 PubMed.
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jørgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Müller F, Forrest AR, Carninci P, Rehli M, Sandelin A. An atlas of active enhancers across human cell types and tissues. Nature. 2014 Mar 27;507(7493):455-461. PubMed.
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014 Jan;42(Database issue):D1001-6. Epub 2013 Dec 6 PubMed.
External Citations
Further Reading
Papers
- Uhlén M, Hallström BM, Lindskog C, Mardinoglu A, Pontén F, Nielsen J. Transcriptomics resources of human tissues and organs. Mol Syst Biol. 2016 Apr 4;12(4):862. PubMed.
Primary Papers
- Dong X, Liao Z, Gritsch D, Hadzhiev Y, Bai Y, Locascio JJ, Guennewig B, Liu G, Blauwendraat C, Wang T, Adler CH, Hedreen JC, Faull RL, Frosch MP, Nelson PT, Rizzu P, Cooper AA, Heutink P, Beach TG, Mattick JS, Müller F, Scherzer CR. Enhancers active in dopamine neurons are a primary link between genetic variation and neuropsychiatric disease. Nat Neurosci. 2018 Oct;21(10):1482-1492. Epub 2018 Sep 17 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
National Institute on Aging
Classifying gene-expression patterns and their relationship to genetic variants is a very important way to determine proximate genetic mechanisms arising from genome-wide association studies (GWAS). The use of laser capture microdissection and RNA-seq is an interesting way to generate these types of data sets; while there are methods for single-cell analysis from human brain, such as DRONC-seq, these tend to have a lot of “missingness” in the data and therefore would be less definitive.
Nonetheless, I am puzzled by several conceptual aspects of this paper. First, I don’t understand the focus on dopamine neurons across neuropsychiatric conditions. In PD, it is now very clear that DA neurons are important but not the only cell type affected, and several enrichment analyses have highlighted a substantial contribution from immunological cells, including microglia. The logic seems a bit circular here and I think we should keep an open mind as to potential mechanisms that might include non-neuronal cells as well as multiple neuronal types, especially if the Braak hypothesis is correct and PD starts in the enteric or olfactory systems.
Second, I am not convinced by the inference that the chromosome 17 risk gene is KANSL1. I do agree that the practice of listing nearest gene to the lead SNP on GWAS can lead to a false sense of precision—we often assume that this means we know the gene when in fact, because of co-inheritance of multiple variants, GWAS nominate loci, not genes. Consequentially, a major difficulty with interpretation of GWAS is that it is difficult to infer causality for any specific variant or gene. Looking through the GTEX dataset, one can see a regulatory effect on KANSL1 of the lead SNP, but also a reasonably strong eQTL at MAPT. Therefore, depending on what evidence one might examine one could favor either KANSL1 or MAPT. Further complicating the picture, there is no reason to assume that there is in fact one gene for each locus—some could easily be complex with contributions by multiple co-regulated genes, for example, or a mixture of rare variants and regulatory events.
Make a Comment
To make a comment you must login or register.