Can We All Agree on How to Draw a Hippo(campus)?
Quick Links
Scientists agree that Alzheimer’s disease ravages the hippocampus, making the volume of this small brain region a key marker for AD clinical trials. But as to precisely where the hippocampus starts and stops on a magnetic resonance image, researchers have little consensus. One tracing protocol can delineate a hippocampal volume more than twice as big as another method, said Giovanni Frisoni, of IRCCS Fatebenefratelli in Brescia, Italy, in a presentation at the Alzheimer’s Association International Conference, held 14-19 July 2012 in Vancouver, Canada. The Alzheimer’s Disease Neuroimaging Initiative (ADNI) has standardized how radiologists should take the images. Now, another collaborative team is stepping in where ADNI left off. The collaboration for A Harmonized Protocol for Hippocampal Volumetry, which led a lively discussion at the meeting, is midway toward its goal of developing a hippocampal trace customized for Alzheimer’s. They have consulted experts to combine disparate methods into one grand, unified protocol they hope everyone will use, making it easy to compare data among different research groups.
“Eventually, in the literature, you will be able to compare apples to apples,” said Simon Duchesne of Université Laval in Québec City, Canada, one of the presenters at the 18 July discussion. Duchesne and principal investigators Frisoni and Clifford Jack, of the Mayo Clinic in Rochester, Minnesota, envision a system whereby wannabe hippocampal tracers the world over can study the protocol, hone their skills on training images, and pass a test to obtain certification. The international project is a joint effort between ADNI and the European Alzheimer’s Disease Consortium, and they are using ADNI images as test cases.
There is no shortage of interest in the new protocol. At the Vancouver meeting, in the sixth such discussion, attendance had quadrupled since the first, Frisoni told Alzforum. The talk grew spirited as researchers debated how best to validate the tracing method. “It is critically important to get standardized protocols,” Laurie Ryan, AD clinical trials coordinator at the National Institute on Aging in Bethesda, Maryland, told Alzforum. “Being able to compare across studies is invaluable.”
The challenge is that defining the precise edge where the hippocampus meets other brain tissue is far from straightforward. It is “painstaking” work, Frisoni noted, and the researchers found 40 different ways to do it in the scientific literature. The project leaders based their plan on the 12 most commonly used methods and consulted 16 expert panelists to define the hippocampus based on four main areas.
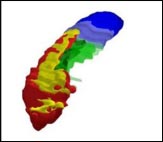
Researchers have defined the most informative hippocampal boundaries for AD, including a minimum hippocampus (red), tail (blue), subiculum (green), and alveus/fimbria (yellow). Image courtesy of Marina Boccardi, Brescia, Italy
The minimum hippocampus (red) is the undisputed hippocampal tissue; every protocol included this section. The tail segment (blue) includes everything behind the minimum hippocampus, which some tracing methods incorporated and others did not. The area called the subiculum (green), similarly, represents differences in how the top 12 protocols delineated the medial edge of the hippocampus. Finally, the researchers included white matter in the form of the alveus/fimbria (yellow), linking the hippocampus to other structures. This white matter is the not “hippocampus proper,” nor is it particularly informative for Alzheimer’s disease, noted project coordinator Marina Boccardi, who also works at the IRCCS in Brescia. However, excluding the white matter completely made it too difficult for tracers to define the hippocampal edge, so they settled on including just the part directly adjacent to the hippocampus.
The resulting hippocampal outline does not directly correspond to the brain’s anatomy. Instead, the researchers wanted to define the region that is most informative, and most reliable to trace, for studies of Alzheimer’s disease. Together, the four regions include all parts of the hippocampus affected by AD.
The current protocol runs 23 pages, with as many figures to illustrate the landmarks that define each region. Using those instructions, Boccardi’s team of five “master tracers” has outlined hippocampi with a correlation of 0.95 among them. This is an “almost perfect” match, Frisoni and Boccardi chorused in a discussion with Alzforum. Such accordance has never before been achieved with manual tracing, Frisoni said; a correlation of 0.8-0.9 is typically considered acceptable.
The next step, Boccardi said, is to validate the evolving protocol with more tracers. The protocol and images are also available to beta users who submit proposals to collaborate with the project. The team hopes to release the final version of the protocol in a year. Not only will the instructions help human tracers align their efforts, but also, researchers writing automated tracing algorithms can take advantage. Frisoni expects the protocol will be built into computerized tracing; this would be the project’s most important utility, he told Alzforum.
Scientists will be able to trace new images according to the standard, as well as go back and retrace old scans to match. With all tracers chiming in on the same tune, researchers should be better able to compare hippocampal volume across any set of drug trials, multiplying the data available to study the disease and medications’ effects.—Amber Dance.
References
External Citations
Further Reading
Papers
- Boccardi M, Ganzola R, Bocchetta M, Pievani M, Redolfi A, Bartzokis G, Camicioli R, Csernansky JG, de Leon MJ, Detoledo-Morrell L, Killiany RJ, Lehéricy S, Pantel J, Pruessner JC, Soininen H, Watson C, Duchesne S, Jack CR, Frisoni GB. Survey of protocols for the manual segmentation of the hippocampus: preparatory steps towards a joint EADC-ADNI harmonized protocol. J Alzheimers Dis. 2011;26 Suppl 3:61-75. PubMed.
- Jack CR, Barkhof F, Bernstein MA, Cantillon M, Cole PE, Decarli C, Dubois B, Duchesne S, Fox NC, Frisoni GB, Hampel H, Hill DL, Johnson K, Mangin JF, Scheltens P, Schwarz AJ, Sperling R, Suhy J, Thompson PM, Weiner M, Foster NL. Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criterion for Alzheimer's disease. Alzheimers Dement. 2011 Jul;7(4):474-485.e4. PubMed.
- Frisoni GB, Jack CR. Harmonization of magnetic resonance-based manual hippocampal segmentation: a mandatory step for wide clinical use. Alzheimers Dement. 2011 Mar;7(2):171-4. PubMed.
News
- The EMA Deems Brain Atrophy Valid Trial Selection Measure
- Paris: Standardization a Hurdle for Spinal Fluid, Imaging Markers
- San Diego: Pre-AD Brain Changes May Allow Early Diagnosis
- Experimental α7 Agonist Meets Cognitive and Clinical Endpoints
- New Assays for Aβ Oligomers in CSF Claim Femtogram Sensitivity
- Q&A With Roche’s CNS Leader Luca Santarelli
- When Is a C9ORF72 Repeat Expansion Not a C9ORF72 Repeat Expansion?
- Wave of New BACE Inhibitors Heading to Phase 2
- Researchers Join to Draw Posterior Cortical Atrophy Out of Shadows
- CSF Markers: Goodbye, Research Use Only; Hello, Clinical
- Metrology, Certification Heavies Take CSF Tests Under Their Wings
- Cochrane Asks for Field’s Input on Draft Reporting Standards
- Aβ’s Good Side—Can It Improve Symptoms of Multiple Sclerosis?
Annotate
To make an annotation you must Login or Register.

Comments
GE Healthcare
The new diagnostic criteria emphasize the use of biomarkers. One important marker of neuronal injury is hippocampal volume as measured by MRI. However, for this marker to become widely used in clinical routine, two things are necessary: 1) the measure must be standardized so one number in one place means the same as the same number measured in another place; and 2) fully automated methods for quantification are necessary.
A necessary step for development of an automated method that uses a standardized protocol is, of course, that the scientific community agrees on such a protocol. This is also where I see the importance of the Frisoni/Jack project—it is a great approach for getting such a consensus around a segmentation protocol. In addition, the project will generate hippocampal masks that will be used for development of automated methods. The project will also generate gold standard data against which automated methods can be validated.
Having measured thousands of hippocampi, allow me to disagree.
Pro primum, I see no need to harmonize hippocampal volumes. Since 1988, when hippocampal atrophy in AD was first presented, there has been no need for such harmony. Hundreds of studies have been published, without harmonization, and a great deal of those studies are perfectly good. Pro secundum, creating a universal hippocampal size is doomed to fail from the very beginning.
Picture this—if I needed to draw hippocampal volumes, I would not compare the volumes from a control pool of mine, or such, or use prior volumes as a reference volume. I would measure patients and controls at the same time. This is because estimates of hippocampal volumes change even among raters over time. The first hippocampal volumes I measured—way back when—were quite a bit larger than later ones. Volumes need to be measured by one person and one person only. Even for a given rater, controls may need to be retraced after a certain period of time, because the volumes may be dynamic. That is, comparing hippocampal volumes to those one drew, say, a year ago, introduces error. In fact, in my opinion, there is no harmony even with one rater. If hippocampal voumes I drew at time 1 and time 2 are out of tune, i.e., not in harmony, how can there be harmony among two or more raters? The hippocampal volume measured by two or more raters results in a fatal error.
Also, there can be no universal hippocampal volume, because the hippocampal size is subject to the size of a person. By this token, the Dutch, who are the tallest population in the world, should have the largest volumes. Eastern races, being shorter, should have smaller volumes. Women—being shorter than most men—have smaller volumes. Then there are a number of other factors to consider: the usual drift, the scanner, the imaging parametres, the alignment, the software used for imaging—i.e., the measuree, the hardware, the software—and in the end, the rater. And you even need to standardize the measurees, given that we know that at least apolipoprotein E alleles and other known and unknown genetic factors influence the volumes. In a manner of speaking, there is too much noise in the concept of harmonized volumes to provide a signal.
In addition, I believe fully automated methods to measure the hippocampus are not science; they are science fiction. Such is the nature of hippocampal structure that it needs a human rater. A machine won't probably ever detect the hippocampal boundaries. That'll be the day when it does.
Some volumes from the literature (right hippocampus) show that measures are highly variable:
Bhatia et al., 1993: 3,770 +/- 610; Bremner et al., 1995: 1,286 +/- 175; Hasboun et al., 1996: 3,420 +/- 490 to 4,180 +/- 530 (depending on alignment); Reiman et al., 1998: 2,462 +/- 347 to 2,519 +/- 454 (depending on ApoE genotype); Watson et al., 1992: 5,263 +/- 652.
The numbers above display almost a fivefold difference in the volumes, and I'd rate all of them acceptable studies. The study by Watson et al., wich has been cited the most, provides volumes which, in my opinion, are twice their actual size. In the end, what really matters is the outcome of the study. In terms of memory impairment, most often it is either conversion to, or the diagnosis of, dementia. The actual numbers have not the slightest interest value. It is the hit rate which counts.
Finally, it takes less than 10 minutes to measure the hippocampus bilaterally. Maybe a tad more if you have to realign or fine-tune some parameters. Some software may be easier to operate than others. Some computers are faster than others. But there really is no trick to manually grade the hippocampus. So why make a very simple issue so hard? Harmonized volumes don't matter. Several studies have provided a ”harmonized” outcome; that, and only the outcomes of the studies, are what matter.
References:
Bhatia S, Bookheimer SY, Gaillard WD, Theodore WH. Measurement of whole temporal lobe and hippocampus for MR volumetry: normative data. Neurology. 1993 Oct;43(10):2006-10. PubMed.
Bremner JD, Randall P, Scott TM, Bronen RA, Seibyl JP, Southwick SM, Delaney RC, McCarthy G, Charney DS, Innis RB. MRI-based measurement of hippocampal volume in patients with combat-related posttraumatic stress disorder. Am J Psychiatry. 1995 Jul;152(7):973-81. PubMed.
Hasboun D, Chantôme M, Zouaoui A, Sahel M, Deladoeuille M, Sourour N, Duyme M, Baulac M, Marsault C, Dormont D. MR determination of hippocampal volume: comparison of three methods. AJNR Am J Neuroradiol. 1996 Jun-Jul;17(6):1091-8. PubMed.
Reiman EM, Uecker A, Caselli RJ, Lewis S, Bandy D, de Leon MJ, De Santi S, Convit A, Osborne D, Weaver A, Thibodeau SN. Hippocampal volumes in cognitively normal persons at genetic risk for Alzheimer's disease. Ann Neurol. 1998 Aug;44(2):288-91. PubMed.
Watson C, Andermann F, Gloor P, Jones-Gotman M, Peters T, Evans A, Olivier A, Melanson D, Leroux G. Anatomic basis of amygdaloid and hippocampal volume measurement by magnetic resonance imaging. Neurology. 1992 Sep;42(9):1743-50. PubMed.
Make a Comment
To make a comment you must login or register.