Danger, S-Bends! New Structure for Aβ42 Fibrils Comes into View
Quick Links
“S” stands for “surprise” in the first atomic-level structure of an Aβ42 fibril. As unveiled on May 4 in Nature Structural and Molecular Biology, each Aβ42 molecule within the fibril formed an S-shaped configuration comprising three β-sheets connected by two loop regions. The new Aβ42 fibril structure differs from what has been reported for its shorter and more innocuous cousin, Aβ40. The researchers, led by Yoshitaka Ishii at the University of Illinois in Chicago, proposed that a salt bridge formed by the last two amino acids in Aβ42 may dictate the structural differences between the two peptides. This could explain why Aβ42 has a higher propensity to aggregate, Ishii believes. It remains uncertain whether other Aβ peptides can take a similar shape. Indeed, the researchers reported in the May 4 Journal of the American Chemical Association that an oligomer structure took a different form.
“Many assume that the structures of Aβ40 and Aβ42 fibrils are quite similar, but our findings suggest that is not the case,” Ishii told Alzforum. He identified the Aβ42 structure based on fibrils made in vitro. It is not clear whether the serpentine configuration represents the form amyloid takes in the Alzheimer’s disease brain. Ishii plans to tackle this question next.
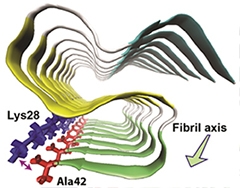
Snaky Fibril.
Aβ42 molecules stack together in a fibril. Each forms an S-shaped structure consisting of three β-strands (blue, yellow, and green) connected by two loop regions (white). A salt bridge between Ala42 and Lys28 stabilizes the structure. [Image courtesy of Xiao et al., Nature Structural and Molecular Biology, 2015.]
Aβ40 and Aβ42 are both cleavage products of the amyloid precursor protein (APP), produced by β-secretase, and Aβ40 predominates in the normal brain. However, the ratio of Aβ42 to Aβ40 rises during AD, and Aβ42 becomes the principal component of amyloid plaques. Researchers have long wondered why Aβ42 aggregates more readily than its shorter counterpart. Because that very propensity has made Aβ42’s structure difficult to pin down, some researchers have instead analyzed fibrils of Aβ40 made in vitro. In its fibrillar form, the Aβ40 monomer reportedly forms a U-shaped structure composed of two β-strands connected by a hairpin loop (see Petkova et al., 2006; Bertini et al., 2011). The U's stack tightly on top of each other to form long strands that can generate fibrils with two- or threefold symmetry. These structure of these fibrils can vary between AD patients (see Parvastu et al., 2008; Sep 2013 news). Some studies identified a roughly similar U-shaped motif for Aβ42 in fibrils; however, a consensus structure did not emerge (see Nov 2005 news; Masuda et al., 2009; Schmidt et al., 2009).
For the current study, first author Yiling Xiao and colleagues used solid-state nuclear magnetic resonance (ssNMR) to home in on the Aβ42 fibril structure. First, the researchers optimized their fibril synthesis and purification protocol. They gently agitated a solution of purified Aβ42 monomers for several days to promote aggregation, then they sonicated the larger clumps of protein that formed to split them into smaller "seeds." Next they added the seeds to a new solution of monomers. They repeated this seeding cycle for four generations, ultimately obtaining homogenous filaments that could be detected by electron microscopy. The researchers prepared the Aβ42 with 13C and 15N amino acids, which facilitated ssNMR.
The initial NMR analysis revealed a strikingly homogeneous structure, as only one set of magnetic resonances appeared for each Aβ42 residue even when the researchers prepared multiple samples. However, when the researchers looked at Aβ42 fibrils after a single round of agitation, without the seeding, they found a variety of structures, suggesting that the seeding cycles selected for a single propagating strain of Aβ42.
The predicted structure based on the NMR spectra consisted of three in-register β-strand regions connected by two loops. Long-range NMR measurements revealed intramolecular contacts between the residues. Computational molecular dynamics modeling, which estimates the movement of atoms, suggested an S-shaped configuration was the most likely structure. A key stabilizer was a salt bridge formed between lysine 28 and the peptide’s final residue, alanine 42. The researchers proposed that this bridge is a defining aspect of Aβ42 structure compared with that of Aβ40. The researchers also identified residues that connect one Aβ42 molecule to its neighbor in the fibril chain: They are glycine 29 and isoleucine 41.
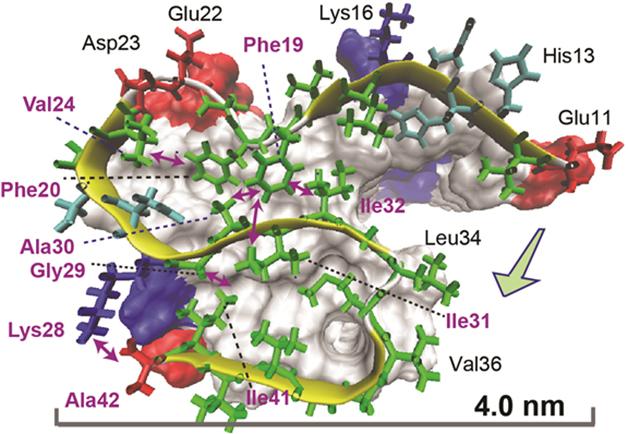
Bonds That Tie. An atomic view of an Aβ42 molecule within a fiber reveals intramolecular connections (double-headed arrows) between residues in different parts of the S-shaped structure. [Image courtesy of Xiao et al., Nature Structural and Molecular Biology, 2015.]
This triple-β-sheet structure differs from the U-shape reported for Aβ40. Would this difference prevent Aβ42 fibrils from seeding Aβ40 monomers and explain why the longer peptide predominates in plaques? To find out, the researchers mixed Aβ42 fibrils with Aβ40. Monitoring fibril growth by uptake of thioflavin-T, they found that Aβ40 incorporated into fibrils after a 13-hour lag phase. However, Aβ40 monomers alone formed fibrils with a similar time course, indicating that the Aβ42 seeds had virtually no seeding sway over Aβ40 fibrillization. In contrast, when the researchers used Aβ40 fibrils as seeds for Aβ40 monomers, fibrils formed immediately.
David Teplow of the University of California, Los Angeles, was enthused about Ishii’s S-shaped structure, in part because it replicated an important element of an Aβ42 structure his lab had reported before. Teplow found that a key difference between Aβ40 and Aβ42 monomers was the existence of a hairpin turn at valine 36 in Aβ42, which is the exact same residue implicated in the second loop of Ishii’s S-shaped Aβ42 fibril (see Roychaudhuri et al., 2013).
How does this Aβ42 fibril structure relate to that of oligomers, which some researchers believe to be the most toxic Aβ species? Ishii said his group has only modeled the S-shaped structure down to the level of a dodecamer, but it is possible that smaller fibrils or oligomers could form the same shape. However, in the JACS paper, Ishii, co-senior author Minako Hoshi of Kyoto University, and colleagues reported the detailed structure of a spherical form of amyloid oligomer that they had previously found to be toxic to neurons and also present in AD brain tissue (see Hoshi et al., 2003; Noguchi et al., 2009). In the new study, the researchers generated these “amylospheroids”—which appear like round blobs under the electron microscope—in vitro. Antibodies they generated to bind to this specific conformation of Aβ suggested that the oligomers were similar to those from the human brain. Some commentators were skeptical about the bioactivity of these oligomers, or whether they represented species that were toxic in the brain. According to ssNMR analysis by first author Sudhakar Parthasarathy, the spheroids were remarkably ordered, consisting of Aβ42 molecules that took on a single U-shaped form similar to that reported for Aβ40 fibrils.
Do different structures reported for monomers, oligomers, and fibrils contradict each other? Not necessarily, according to Teplow, who collaborated on earlier studies that characterized the amylospheroids. “Aβ is intrinsically disordered,” Teplow said. Thus, multiple structures are possible and can be swayed by oligomerization or fibrillization.
Brigita Urbanc of Drexel University in Philadelphia, who uses computation to work out the structure of Aβ42 oligomers, noted that previous work from her lab had predicted that the C-terminal Ala42 residue played a key role in dictating oligomer structure, much as it does in Ishii’s S-bend fibril structure (see Urbanc et al., 2004; Urbanc et al., 2010).
Ishii said he plans to characterize fibrils from patients to see how they compare to the S-shaped structure. Until he does, it is difficult to know how closely the structure mimics what occurs in the brain. He acknowledged that multiple strains of Aβ42 fibril likely exist. He added that the stability of the S-shaped structure, if it exists in the brain, likely allows for efficient fibril formation there. Antibodies targeting this structure—for example, the salt bridge region—could theoretically remove fibrils and stop their formation, Ishii said. Furthermore, he believes such fibrils act as a reservoir of oligomers, so ridding the brain of fibrils could take a toll on oligomers as well, he said.
Robert Tycko of the National Institutes of Health in Bethesda, Maryland, who characterized the U-shaped structure of Aβ40 fibrils, was struck by how much Ishii’s Aβ42 structure differed. He also suspects the situation could be different in the brain. “In general, Aβ fibrils are polymorphic, meaning that the same peptides can form multiple types of fibril with different internal molecular structures,” he wrote. “In future studies, it will be important to characterize structures of Aβ42 fibrils that develop in human brain tissue.”
Beyond determining whether the structures even exist in the brain, researchers will need to determine which ones matter, commented Charles Glabe of the University of California, Irvine. “The challenge for the future will be understanding which structures are more closely related to pathology and whether they are pathogenic by the same mechanism.”—Jessica Shugart
References
News Citations
- Does Aβ Come In Strains? Glimpse Into Human Brain Suggests Yes
- See How They Grow: Structure of Amyloid-β Fibrils
Paper Citations
- Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer's beta-amyloid fibrils. Biochemistry. 2006 Jan 17;45(2):498-512. PubMed.
- Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi A. A new structural model of Aβ40 fibrils. J Am Chem Soc. 2011 Oct 12;133(40):16013-22. PubMed.
- Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2008 Nov 25;105(47):18349-54. PubMed.
- Masuda Y, Uemura S, Ohashi R, Nakanishi A, Takegoshi K, Shimizu T, Shirasawa T, Irie K. Identification of physiological and toxic conformations in Abeta42 aggregates. Chembiochem. 2009 Jan 26;10(2):287-95. PubMed.
- Schmidt M, Sachse C, Richter W, Xu C, Fändrich M, Grigorieff N. Comparison of Alzheimer Abeta(1-40) and Abeta(1-42) amyloid fibrils reveals similar protofilament structures. Proc Natl Acad Sci U S A. 2009 Nov 24;106(47):19813-8. PubMed.
- Roychaudhuri R, Yang M, Deshpande A, Cole GM, Frautschy S, Lomakin A, Benedek GB, Teplow DB. C-Terminal Turn Stability Determines Assembly Differences between Aβ40 and Aβ42. J Mol Biol. 2013 Jan 23;425(2):292-308. PubMed.
- Hoshi M, Sato M, Matsumoto S, Noguchi A, Yasutake K, Yoshida N, Sato K. Spherical aggregates of beta-amyloid (amylospheroid) show high neurotoxicity and activate tau protein kinase I/glycogen synthase kinase-3beta. Proc Natl Acad Sci U S A. 2003 May 27;100(11):6370-5. PubMed.
- Noguchi A, Matsumura S, Dezawa M, Tada M, Yanazawa M, Ito A, Akioka M, Kikuchi S, Sato M, Ideno S, Noda M, Fukunari A, Muramatsu S, Itokazu Y, Sato K, Takahashi H, Teplow DB, Nabeshima Y, Kakita A, Imahori K, Hoshi M. Isolation and characterization of patient-derived, toxic, high mass amyloid beta-protein (Abeta) assembly from Alzheimer disease brains. J Biol Chem. 2009 Nov 20;284(47):32895-905. PubMed.
- Urbanc B, Cruz L, Yun S, Buldyrev SV, Bitan G, Teplow DB, Stanley HE. In silico study of amyloid beta-protein folding and oligomerization. Proc Natl Acad Sci U S A. 2004 Dec 14;101(50):17345-50. PubMed.
- Urbanc B, Betnel M, Cruz L, Bitan G, Teplow DB. Elucidation of amyloid beta-protein oligomerization mechanisms: discrete molecular dynamics study. J Am Chem Soc. 2010 Mar 31;132(12):4266-80. PubMed.
Further Reading
Primary Papers
- Xiao Y, Ma B, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y. Aβ(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease. Nat Struct Mol Biol. 2015 Jun;22(6):499-505. Epub 2015 May 4 PubMed.
- Parthasarathy S, Inoue M, Xiao Y, Matsumura Y, Nabeshima Y, Hoshi M, Ishii Y. Structural Insight into an Alzheimer's Brain-Derived Spherical Assembly of Amyloid β by Solid-State NMR. J Am Chem Soc. 2015 May 27;137(20):6480-3. Epub 2015 May 18 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
NIDDK, NIH
This structural model for Aβ42 fibrils is surprisingly different from existing models for Aβ40 fibrils. The peptide backbone conformation is different, and the contacts between amino acid side chains are different. For example, most Aβ40 fibrils have contacts between side chains of Phe19 and Leu34. Some Aβ40 fibrils have contacts between side chains of Asp23 and Lys28. These contacts are not present in the Aβ42 fibrils examined by Ishii and coworkers. As they suggest, these pronounced differences in backbone conformation and side chain contacts may explain the lack of efficient cross-seeding between Aβ42 fibrils and Aβ40 fibrils.
In general, Aβ fibrils are polymorphic, meaning that the same peptides can form multiple types of fibrils with different internal molecular structures. It is still possible that certain Aβ42 fibril structures have structures that are more similar to those of Aβ40 fibrils, and that certain Aβ42 fibrils are indeed capable of seeding the growth of Aβ40 fibrils. In future studies, it will be important to characterize structures of Aβ42 fibrils that develop in human brain tissue (as opposed to fibrils grown in vitro). In 2013, my group published a structural model for Aβ40 fibrils from brain tissue (Lu et al., 2013), but so far this has not been done for Aβ42 fibrils. It will be interesting to see how different the brain-derived fibril structures are.
References:
Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular Structure of β-Amyloid Fibrils in Alzheimer's Disease Brain Tissue. Cell. 2013 Sep 12;154(6):1257-68. PubMed.
View all comments by Robert TyckoUniversity of California, Irvine
This is an interesting paper that underscores the conformational polymorphisms in β-sheet aggregates of Aβ. This one is unique in two aspects: It is the first structure that is a triple parallel, in-register sheet, and it is unique to Aβ42. I’m not sure how many unique structures there are so far, but at least four distinct, parallel, in-register structures. There is also evidence for antiparallel β-sheet oligomers, but the structural details of these are not as well-known as the parallel β-sheets.
The triple-sheet 42 structure does not appear to seed 40 fibrillization, so there appears to be at least two structures that Aβ42 can adopt: the unique triple-sheet structure that does not seed Aβ40 and a structure that can seed Aβ40 fibrillization that may be a two-sheet β hairpin (Jarrett et al., 1993). The unique structure of Aβ42 is consistent with results obtained with several conformation-dependent monoclonal antibodies that appear to be specific for Aβ42 fibrils and do not recognize Aβ40 fibrils, but yet recognize amino terminal and central epitopes rather than the carboxyl terminus (Hatami et al., 2014). The challenge for the future will be understanding which structures are more closely related to pathology and whether they are pathogenic by the same mechanism.
References:
Jarrett JT, Berger EP, Lansbury PT. The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer's disease. Biochemistry. 1993 May 11;32(18):4693-7. PubMed.
Hatami A, Albay R 3rd, Monjazeb S, Milton S, Glabe C. Monoclonal antibodies against Aβ42 fibrils distinguish multiple aggregation state polymorphisms in vitro and in Alzheimer disease brain. J Biol Chem. 2014 Nov 14;289(46):32131-43. Epub 2014 Oct 3 PubMed.
View all comments by Charles GlabeUCLA Neuropsychiatric Institute
The authors describe the results of a solid-state NMR study on amylospheroid (ASPD), a toxic Aβ1-42 oligomer. They demonstrate the similarity of the synthetic ASPD to native ASPD and go on to suggest that the secondary structure of this particle is largely parallel β sheet. They further suggest this method may be suitable to study other toxic amyloid oligomers. Understanding the structure of toxic Aβ oligomers is a hot topic in the field, as we all hope to understand pathogenicity through structural knowledge. This has been difficult to date due to the technical problems of Aβ oligomer assembly and structural determination of polymorphic complexes. Repetition of this work by others will be critical to defining its usefulness. The suggestion that β sheet is a prominent structure within the particle is not unexpected, but has profound implications for toxicity. The tendency of β sheet structures to be pore-forming peptides is well established, and these data lend further weight to the notion that pore formation may play a role in amyloid toxicity. A major obstacle to further work is the fact that the relevant toxic structures may form in a membrane environment that may differ substantially from the aqueous phase. Still, this research establishes a method for structure determination that is quite powerful and could lead to significant advances. Since so many toxic oligomer species have been described, caution is warranted before we crown any particular one as the "pathogenic species."
View all comments by Bruce KaganMake a Comment
To make a comment you must login or register.