Affibodies—Putting the β in Aβ?
Quick Links
Aβ is infamous for its ability to form large fibrils rich in β-sheet secondary structure. In contrast, soluble monomeric Aβ is totally different and thought to assume random or α-helical structures. But in this week’s PNAS, researchers led by Torleif Härd at the University of Goteborg, Sweden, debut a monomeric Aβ structure that has a β-sheet hairpin loop. The structure is courtesy of a protein ligand, or affibody, that has high affinity for Aβ monomers. This ligand, called ZAβ3, surrounds Aβ1-40, stabilizing the β-sheets. The latter are uncannily similar, though not identical, to the β-sheet hairpin loops of amyloid fibrils. “The discovery of this hairpin leads to possible mechanisms for oligomer and fibril formation, and the binder itself is very exciting because it has practical uses in research and also as a basis for therapy,” said Härd in an interview with ARF. Whether this structure occurs alone in solution, much less in vivo, or is partly driven by the affibody will need to be tested in future studies.
Affibodies were engineered at the Royal Institute of Technology, Stockholm, and are now patented and developed by Affibody AB, Bromma, Sweden. They are derived from the Z domain of staphylococcal protein A, which binds avidly to the Fc stub of immunoglobulin G. Two helices of the Z domain can be randomly changed to generate a wide range of binding surfaces, much like the variable chains on antibodies. For example, some affibodies bind with high affinity to insulin, TNFα, and ErbB2, which is overexpressed in some breast cancer cells. Last year researchers at Stockholm University reported that they had generated affibodies to Aβ (see Grönwall et al., 2007), and it is these ligands that Härd and colleagues studied at molecular resolution.
First author Wolfgang Hoyer and colleagues characterized the 16 Aβ affibodies that the group in Stockholm had generated. They report that all of them need a cysteine residue at amino acid 28 (each affibody has 58 amino acids) for high-affinity binding, suggesting that the affibodies act as dimers held together by disulphide bonds. Binding stoichiometry supports that idea. A dimer of ZAβ3 binds 1:1 with Aβ at nanomolar affinity. NMR data support this structure, too. The researchers obtained high-precision NMR spectra that show, at atomic resolution, Aβ buried in a cavity formed by a ZAβ3 dimer. Residues 17-36 of Aβ form a β-sheet hairpin loop that is stabilized by the hydrophobic core of that cavity (see figure below).
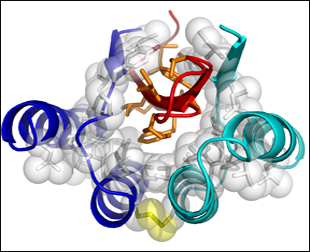
High-precision NMR Structure of Aβ-ZAβ3
A dimer of ZAβ3 (blue, turquoise), linked by a disulphide bond (yellow), forms a high-affinity complex with Aβ1-40. Residues 17-36 of Aβ (red) form a β-turn-β hairpin loop in the hydrophobic core of the complex. Aβ hydrophobic side chains exposed to water are shown in orange. Image credit: National Academy of Sciences, PNAS © 2008
The authors write that this is the first high-resolution structure of soluble Aβ in β conformation. Previous structural determinations of Aβ monomers have been based on only portions of the molecule (see ARF related news story), suggest random coil or helical structures (see, e.g., Miles et al., 2008), or have been obtained in lipid micelles (see, e.g., Rangachari et al., 2007). “This is an interesting structure of another monomeric conformation. It has some resemblance to the fibril structure in that the same regions that form extended strands in the fibril also form β strands in this monomer,” said Charlie Glabe, University of California, Irvine, in an interview with ARF. (Glabe and colleagues are pursuing antibody-based strategies to stabilize and study Aβ oligomers.)
The difference is that in this structure, the β strands bind each other within the Aβ molecule, whereas in fibrils they bind strands on different Aβ molecules. “This is what we think people might find interesting,” suggested Härd, and in fact the authors used this difference as a basis for proposing a model of aggregation. Their model predicts that Aβ monomers, with intramolecularly bound β-sheets, layer upon one another before undergoing a concerted 90-degree rotation around their strand axes (much like closing Venetian blinds), which would effectively switch the bonding from intra- to intermolecular. Glabe questioned whether this conformation change could work without dissociation. “Basically, every strand in the soluble oligomer intermediate would have to rotate 90 degrees coordinately, in the same direction, at the same time,” he said.
It is also not clear if the Aβ β-sheet structure would form in the absence of the ZAβ3 ligand, given that such an Aβ conformation would expose a considerable amount of hydrophobic side chains to the solvent, though there is evidence it might. In phage display, which is how the researchers isolated the affibodies, this β structure seems well represented. “We find that when you select for binding, in 16 cases you select a very well-defined hairpin structure of Aβ, so that structure is available to the monomer,” suggested Härd. And the affibody may explain why others have not seen a similar monomeric structure. “If you look at antibodies, they usually bind to several different aggregates and not just the monomer, or they bind just to the N-terminus, so I think it is this affibody that lets us see Aβ in this confirmation so nicely,” added Hoyer.
Affibodies are currently being used for diagnostic purposes and as a potential cancer therapy. One other interesting property of the affibody is that it is a potent inhibitor of aggregation. It is unclear whether Aβ affibodies might make useful therapeutics for AD. They do not readily cross the blood-brain barrier and would likely be immunogenic, but Härd and colleagues are exploring if they can ameliorate toxicity in fruit fly models of AD. “From a biotech point of view, it really adds another example of protein engineering as an alternative to immunotherapy or antibody-based methods. There are many tracks this research could take, but the most important is to bring this whole system into in vivo conditions to see what can be found,” said Härd.—Tom Fagan
References
News Citations
Paper Citations
- Grönwall C, Jonsson A, Lindström S, Gunneriusson E, Ståhl S, Herne N. Selection and characterization of Affibody ligands binding to Alzheimer amyloid beta peptides. J Biotechnol. 2007 Jan 30;128(1):162-83. PubMed.
- Miles LA, Wun KS, Crespi GA, Fodero-Tavoletti MT, Galatis D, Bagley CJ, Beyreuther K, Masters CL, Cappai R, McKinstry WJ, Barnham KJ, Parker MW. Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope. J Mol Biol. 2008 Mar 14;377(1):181-92. PubMed.
- Rangachari V, Moore BD, Reed DK, Sonoda LK, Bridges AW, Conboy E, Hartigan D, Rosenberry TL. Amyloid-beta(1-42) rapidly forms protofibrils and oligomers by distinct pathways in low concentrations of sodium dodecylsulfate. Biochemistry. 2007 Oct 30;46(43):12451-62. PubMed.
Further Reading
Papers
- Khandogin J, Brooks CL. Linking folding with aggregation in Alzheimer's beta-amyloid peptides. Proc Natl Acad Sci U S A. 2007 Oct 23;104(43):16880-5. PubMed.
- Gardberg AS, Dice LT, Ou S, Rich RL, Helmbrecht E, Ko J, Wetzel R, Myszka DG, Patterson PH, Dealwis C. Molecular basis for passive immunotherapy of Alzheimer's disease. Proc Natl Acad Sci U S A. 2007 Oct 2;104(40):15659-64. PubMed.
Primary Papers
- Hoyer W, Grönwall C, Jonsson A, Ståhl S, Härd T. Stabilization of a beta-hairpin in monomeric Alzheimer's amyloid-beta peptide inhibits amyloid formation. Proc Natl Acad Sci U S A. 2008 Apr 1;105(13):5099-104. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
�
Hoyer and coworkers have solved a structure of the amyloid-β peptide in complex with a phage-display selected affibody using NMR spectroscopy. The affibody is responsible for stabilizing the Aβ monomer by inhibiting fibril formation. The Aβ adopts a parallel β-hairpin structure, where the two β-strands consisting of residues 15-22 (strand A) and 30-36 (strand B) form intramolecular hydrogen bonds between each other. Strand A is stabilized by a short strand from the affibody which runs anti-parallel, while strand B is stabilized by a short strand that is parallel to it.
From a large body of data, we know that fibrils exhibit a “cross-β” pattern in x-ray fiber diffraction (1). This is associated with a fundamental structure consisting of extended β-sheet networks in which peptide chains are displayed perpendicular to the fibril axis, while the hydrogen bonding direction of the sheet is parallel to the fibril axis (2,3). Hence, the direction of the hydrogen bonds of a conventional β-hairpin as observed in the current study will not fit the bill.
The authors acknowledge this fact, and propose that what they observe might be an intermediate structure. In fact, the authors suggest that their structure may amount to the toxic soluble oligomers associated with AD. If this is true, the strands A and B will have to rotate 90 degrees about their axis in order to form the in-register parallel β structure that is likely to form the fibrils. While one cannot rule out this possibility, one must be cautious with such interpretations. It is equally possible that the affibody might be responsible for forming a stable, mixed β-sheet structure. In any event, the affibodies can be useful for passive immunotherapy via the peripheral sink theory (4), as they bind Aβ monomers. Hence, this is an interesting study and the main conclusions require further validation.
References:
Sunde M, Blake C. The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv Protein Chem. 1997;50:123-59. PubMed.
Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005 Jan 14;307(5707):262-5. PubMed.
Guo JT, Wetzel R, Xu Y. Molecular modeling of the core of Abeta amyloid fibrils. Proteins. 2004 Nov 1;57(2):357-64. PubMed.
Solomon B. Intravenous immunoglobulin and Alzheimer's disease immunotherapy. Curr Opin Mol Ther. 2007 Feb;9(1):79-85. PubMed.
Drexel University
Capturing Aβ Using Engineered Affinity Proteins
Alzheimer disease (AD) is associated with the amyloid-β protein (Aβ) which assembles into toxic oligomers, protofibrils, and fibrils, and is the major component of amyloid plaques in the AD brain. Substantial evidence implicates the early stages of Aβ assembly in the onset of the disease. Many different strategies that aim at preventing Aβ molecules from formation of toxic assemblies are currently under investigation.
The present study by Hoyer et al. was motivated by novel therapeutic strategies that explore ways to create a peripheral sink mechanism by administering an Aβ binding molecule, a ligand, with the capacity to reduce Aβ in the central nervous system by channeling it into the plasma. As the Aβ1-40 binding molecule, Hoyer et al. proposed to apply an engineered affinity protein (affibody), ZAβ3, based on the Z domain derived from the staphylococcal protein A. In their paper, Hoyer et al. presented 16 different ligands which were previously shown to bind both Aβ1-40 and Aβ1-42 and to form dimers through the intermolecular disulfide bonding between Cys28. Using a combination of powerful methods—isothermal titration calorimetry, circular dichroism, and heteronuclear single quantum correlation (HSQC) NMR spectroscopy—Hoyer et al. derived detailed structural information on the observed ZAβ3:Aβ1-40 complex.
Dimeric ZAβ3 was shown to bind monomeric Aβ1-40 with 1:1 stoichiometry. Binding of ZAβ3 was coupled to folding of Aβ1-40 and the ligand itself. Aβ1-40 was shown to undergo a conformational transition into a β-hairpin conformation upon binding. ZAβ3 dimer, composed of two β-strands and four α-helices wrapping around the Aβ1-40 monomer, created a large hydrophobic tunnel-like cavity. Aβ1-40 within this cavity adopted a β-hairpin with two strands, 17-23 and 30-36, connected by intramolecular backbone hydrogen bonds. Importantly, the cavity of ZAβ3 dimer was inaccessible to water, and the nonpolar faces of the Aβ1-40 β-hairpin inside the cavity were thus mostly shielded from the solvent. The N-terminal region of Aβ1-40, 1-15, and the terminal regions 1-13 and 57-58 of both ZAβ3 subunits were not well defined by the NMR constraints, implying lack of any ordered structure in these regions.
The significance of the ZAβ3 ligand as an inhibitor of Aβ1-40 assembly is that it binds to the hydrophobic part of Aβ1-40 and thereby disrupts Aβ’s potential for further β-sheet extension. To make that possible, the ZAβ3 ligand was engineered such that the two solvent-exposed short β-strands of the ZAβ3 dimer have little propensity to form either α-helical or β-sheet structure.
Hoyer et al. discussed the β-hairpin monomer conformation of Aβ1-40 in relation to the conformation of individual Aβ1-40 molecules within a fibril as proposed in a fibril model by Petkova et al. (1). While the hook-like structure of the β-hairpin monomer resembles the hook-like structure in the fibril model, the hydrogen bonding in the latter is rotated by a right angle with respect to the former. Such a rotation requires a significant structural change as it involves the breaking of intramolecular hydrogen bonds (to destabilize the β-hairpin monomer) and the formation of new intermolecular hydrogen bonds between neighboring Aβ1-40 molecules.
Hoyer et al. suggest that the β-hairpin monomer conformation may be a key conformation for fibril formation to take place. Here, I will give some insights on the significance of the β-hairpin Aβ1-40 conformation from the point of view of a computational physicist. Even though the β-hairpin conformation of Aβ1-40 monomer may be accessible, it may not be energetically favorable under normal aqueous conditions. On the contrary, in aqueous solutions Aβ1-40 monomer adopts a collapsed-coil conformation with less that 15 percent of total secondary structure (2,3). The interactions driving the folding into a collapsed-coil conformation are primarily due to the effective hydrophobicity as also corroborated by computational studies (4). The fact that Hoyer et al. observed a β-hairpin monomer structure of Aβ1-40 within the ZAβ3:Aβ1-40 complex is very much to do with the particular local environment associated with Aβ1-40 folding. The fact that the observed cavity formed by the ZAβ3 dimer is water inaccessible is critical for understanding Aβ1-40 β-hairpin formation. With no water present, there is no hydrophobic effect, so Aβ1-40 folding cannot be driven by the hydrophobic collapse as in the case of aqueous environment. In the absence of the hydrophobic effect, molecular dynamics simulations indeed show that a β-hairpin and other β-sheet monomer structures with up to four β-strands are formed, leading to formation of planar β-sheet dimers (5). This same study also showed that in water, these planar β-sheet monomer and dimer conformations would be equally favorable to both Aβ1-40 and Aβ1-42—a result that is difficult to reconcile with the fact that Aβ1-42 forms larger assemblies faster than does Aβ1-40. While the computational study of Aβ oligomer formation indicated the presence of a turn/loop centered at G25-S26 in both Aβ1-40 and Aβ1-42, with the local structure that resembles the β-hairpin, the intramolecular hydrogen bonding is at best weak, consistent with the importance of effective hydrophobicity in oligomer formation (4). Finally, the presence of the intramolecular hydrogen bonding in a β-hairpin conformation may actually increase the free energy barrier for transition from a monomeric β-hairpin into a fibril-like structure because breaking the hydrogen bonds in a β-hairpin is energetically unfavorable. Thus, it would be energetically more favorable to form a fibril-like structure from collapsed-coil monomer/oligomer conformations, which already possess a turn structure at G25-S26, held together by hydrophobically driven forces rather than intramolecular hydrogen bonds.
In conclusion, Aβ1-40 monomer forming the β-hairpin structure upon dimeric ZAβ3 binding may create a large free energy barrier preventing Aβ1-40 to form toxic assemblies and thus provide a new and exciting therapeutic strategy.
References:
Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer's beta -amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002 Dec 24;99(26):16742-7. PubMed.
Zhang S, Iwata K, Lachenmann MJ, Peng JW, Li S, Stimson ER, Lu Y, Felix AM, Maggio JE, Lee JP. The Alzheimer's peptide a beta adopts a collapsed coil structure in water. J Struct Biol. 2000 Jun;130(2-3):130-41. PubMed.
Kirkitadze MD, Condron MM, Teplow DB. Identification and characterization of key kinetic intermediates in amyloid beta-protein fibrillogenesis. J Mol Biol. 2001 Oct 5;312(5):1103-19. PubMed.
Urbanc B, Cruz L, Yun S, Buldyrev SV, Bitan G, Teplow DB, Stanley HE. In silico study of amyloid beta-protein folding and oligomerization. Proc Natl Acad Sci U S A. 2004 Dec 14;101(50):17345-50. PubMed.
Urbanc B, Cruz L, Ding F, Sammond D, Khare S, Buldyrev SV, Stanley HE, Dokholyan NV. Molecular dynamics simulation of amyloid beta dimer formation. Biophys J. 2004 Oct;87(4):2310-21. PubMed.
St Vincent's Institute
Grönwall et al. (2007) recently applied phage-display techniques to identify affibody ligands that efficiently capture Aβ peptides from human plasma and serum. In the present paper by Hoyer et al., one of these phage-selected affibodies, Zab3, is shown to form a disulfide cross-linked dimer that binds monomeric Aβ(1-40) with low nanomolar affinity as determined by isothermal titration calorimetry. In addition, Hoyer et al. used thioflavin T fluorescence to show that the Zab3 dimer completely inhibits Aβ(1-40) fibrillization at stoichiometric concentrations to the Aβ monomer.
As such, Zab3 represents a novel lead molecule for the development of therapeutics to promote amyloid plaque clearance in vivo according to the peripheral sink mechanism of amyloid dissolution. However, the necessity for a disulfide-linked dimer to bind the peptide may limit its development potential. Although the affibody shows promise in inhibiting fibrils, no data were presented as to whether it can break up fibrils once formed or break up Aβ oligomers which are thought to be the toxic species in the disease.
Hoyer et al. also used NMR spectroscopy to investigate the molecular basis for Zab3 recognition of Aβ(1-40), and report the high-resolution complex structure of Aβ (1-40) bound to recombinantly expressed Zab3 in solution. The Aβ conformation determined is a β-hairpin where intramolecular backbone hydrogen bonds are observed between two strands across Aβ(17-23 and 30-36).
This structure represents the first high-resolution structure of the Aβ peptide in a β conformation. The N-terminal region (1-16) of Aβ is disordered in the model. This is consistent with recent structures of antibodies complexed to N-terminal fragments of Aβ (Gardberg et al., 2007; Miles et al., 2008). In our recent structures of Aβ1-16 and 1-28 complexed at the N-terminus to the antibody WO2, the core epitope (2-8) exhibited an extended conformation, while no structure was observed outside of the confines of the antibody binding region (due to flexibility in the residues immediately C-terminal to the epitope).
Importantly, each face of the Aβ-hairpin structure is largely non-polar, and this conformation of Aβ constitutes the hydrophobic core of the complex structure. The question arises of whether this conformation represents a valid intermediate towards fibrillization as considered by the authors, or is indeed an artifact structure accommodated by the hydrophobic cleft of Zab3.
Considering the pleomorphic nature of Aβ peptides and the issues highlighted in this forum around the incongruous hydrogen bonding direction observed in this model and in the fibrillar form, at this stage the latter is probable—but not established. However, this does not diminish the importance of this work in providing a molecular template for recognition of the central and C-terminal regions of Aβ, and in providing insights into the inhibition of fibrillization.
This model is potentially useful in the development of therapeutics designed to clear amyloid plaques via the peripheral sink mechanism. A particularly interesting finding is the similarity of the Zab3 strands that cap the Aβ-hairpin to β-sheet blockers in development as fibrillogenesis inhibitors.
Finally, it will be fascinating to see whether the affibody approach will lead to molecules that can disrupt preformed Aβ oligomers and protofibrils. It has been observed that metal ions can modulate Aβ toxicity and aggregation in neuronal cultures, with Cu2+ ions increasing Aβ toxicity whilst Zn2+ ions attenuate it (Cuajungco et al., 2005). Thus, drugs such as affibodies that target the oxidative pathway in the disease and reduce the production of toxic Aβ peptides constitute a promising therapeutic approach.
References:
Grönwall C, Jonsson A, Lindström S, Gunneriusson E, Ståhl S, Herne N. Selection and characterization of Affibody ligands binding to Alzheimer amyloid beta peptides. J Biotechnol. 2007 Jan 30;128(1):162-83. PubMed.
Gardberg AS, Dice LT, Ou S, Rich RL, Helmbrecht E, Ko J, Wetzel R, Myszka DG, Patterson PH, Dealwis C. Molecular basis for passive immunotherapy of Alzheimer's disease. Proc Natl Acad Sci U S A. 2007 Oct 2;104(40):15659-64. PubMed.
Miles LA, Wun KS, Crespi GA, Fodero-Tavoletti MT, Galatis D, Bagley CJ, Beyreuther K, Masters CL, Cappai R, McKinstry WJ, Barnham KJ, Parker MW. Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope. J Mol Biol. 2008 Mar 14;377(1):181-92. PubMed.
Cuajungco MP, Frederickson CJ, Bush AI. Amyloid-beta metal interaction and metal chelation. Subcell Biochem. 2005;38:235-54. PubMed.
View all comments by Michael ParkerMake a Comment
To make a comment you must login or register.