Mutations
MAPT G86S
Quick Links
Overview
Pathogenicity: Frontotemporal Dementia : Unclear Pathogenicity
Clinical
Phenotype: None
Reference Assembly: GRCh37/hg19
Position: Chr17:44051786 G>A
dbSNP ID: rs63751135
Coding/Non-Coding: Coding
DNA
Change: Substitution
Expected RNA
Consequence: Substitution
Expected Protein
Consequence: Missense
Codon
Change: GGC to AGC
Reference
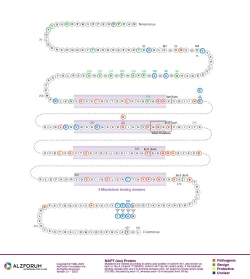
Isoform: Tau Isoform Tau-F (441 aa)
Genomic
Region: Exon 3
Findings
This mutation was identified in a woman diagnosed with frontotemporal dementia with onset at age 71. She presented with decreased concentration and activity, distractibility, perseveration, and personality changes. She later developed frontal release signs and seizures, and became mute and unable to walk without assistance. The proband had a positive family history of neurodegenerative disease, including a mother with unspecified dementia and two siblings with clinical features consistent with Alzheimer's disease. The mutation was not present in either sibling and the mother's DNA was unavailable. The mutation was also absent in 100 control chromosomes, but pathogenicity could not be determined (Stanford et al., 2004).
Neuropathology
Unknown. A PET scan showed frontal hypometabolism (Stanford et al., 2004).
Biological Effect
Exon-trapping experiments indicate that the G86S mutation does not alter the normal splicing of exons 2 or 3. The amino acid substitution results in the creation of phosphorylation and O-glycosylation sites, as predicted (Stanford et al., 2004).
Last Updated: 16 Feb 2023
References
Paper Citations
- Stanford PM, Brooks WS, Teber ET, Hallupp M, McLean C, Halliday GM, Martins RN, Kwok JB, Schofield PR. Frequency of tau mutations in familial and sporadic frontotemporal dementia and other tauopathies. J Neurol. 2004 Sep;251(9):1098-104. PubMed.
Further Reading
Learn More
Protein Diagram
Primary Papers
- Stanford PM, Brooks WS, Teber ET, Hallupp M, McLean C, Halliday GM, Martins RN, Kwok JB, Schofield PR. Frequency of tau mutations in familial and sporadic frontotemporal dementia and other tauopathies. J Neurol. 2004 Sep;251(9):1098-104. PubMed.
Alzpedia
Disclaimer: Alzforum does not provide medical advice. The Content is for informational, educational, research and reference purposes only and is not intended to substitute for professional medical advice, diagnosis or treatment. Always seek advice from a qualified physician or health care professional about any medical concern, and do not disregard professional medical advice because of anything you may read on Alzforum.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.