Mutations
MAPT A90V
Quick Links
Overview
Pathogenicity: Alzheimer's Disease : Unclear Pathogenicity
Clinical
Phenotype: Alzheimer's Disease
Reference Assembly: GRCh37/hg19
Position: Chr17:44051799 C>T
dbSNP ID: NA
Coding/Non-Coding: Coding
DNA
Change: Substitution
Expected RNA
Consequence: Substitution
Expected Protein
Consequence: Missense
Codon
Change: GCC to GTC
Reference
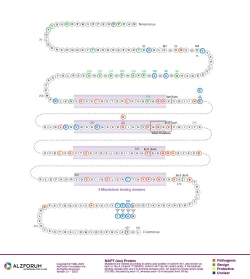
Isoform: Tau Isoform Tau-F (441 aa)
Genomic
Region: Exon 3
Findings
This variant was detected in an individual with apparently sporadic AD (Sala Frigerio et al., 2015). There was no family history of dementia. Other than a diagnosis of AD, clinical details related to this individual were not reported.
Neuropathology
Unknown.
Biological Effect
The pathogenicity of this variant is unclear; it may be simply a rare polymorphism. PolyPhen-2 predicts that it is benign in silico.
Last Updated: 16 Feb 2023
References
Other Citations
Further Reading
No Available Further Reading
Protein Diagram
Primary Papers
- Sala Frigerio C, Lau P, Troakes C, Deramecourt V, Gele P, Van Loo P, Voet T, De Strooper B. On the identification of low allele frequency mosaic mutations in the brains of Alzheimer's disease patients. Alzheimers Dement. 2015 Apr 29; PubMed.
Alzpedia
Disclaimer: Alzforum does not provide medical advice. The Content is for informational, educational, research and reference purposes only and is not intended to substitute for professional medical advice, diagnosis or treatment. Always seek advice from a qualified physician or health care professional about any medical concern, and do not disregard professional medical advice because of anything you may read on Alzforum.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.